INTRODUCTION
Although the clinical features of most foodborne illnesses are non-specific for individual cases, outbreaks frequently have characteristic features with respect to incubation periods, duration of symptoms, and the percentage of cases that experience specific signs and symptoms which depend on the aetiology [Reference Olson1, Reference Hall2]. These clinical profiles have been used to assign likely microbial causes to foodborne outbreaks in the United States for which no pathogen was isolated by routine laboratory testing, and have demonstrated that a high proportion of such outbreaks were consistent with outbreaks of norovirus [Reference Deneen3, Reference Fankhauser4]. Similarly, recognizing that outbreak characteristics are not consistent with norovirus has been used to guide laboratory testing to identify diarrhoeagenic Escherichia coli strains in outbreaks caused by enterotoxigenic E. coli (ETEC), and atypical enteropathogenic E. coli (EPEC) [Reference Dalton5–Reference Beatty7].
In addition to improving diagnosis in the outbreak setting, the prospective application of clinical profiling can facilitate the environmental investigation of outbreaks by providing a context for conducting the investigation, as different pathogens behave differently in the environment. For example, norovirus has a human reservoir and does not replicate on food or in the environment. Thus, identifying norovirus as a likely agent should allow the environmental health specialist (EHS) to focus on issues of food-worker health and hygiene, and the potential for food-workers to cause ongoing transmission. In contrast, the bacteria Bacillus cereus and Staphylococcus aureus do multiply and produce toxins in foods at temperatures that allow them to do so. Thus, identifying opportunities for time and temperature abuse of foods would be indicated if the outbreak appeared to be caused by these diarrhoeal or vomiting toxins. Allowing the EHS to focus the investigation before results of cultures are available enables the investigation to be completed more rapidly, with a greater chance of successfully identifying the underlying cause of the outbreak. For outbreaks in commercial establishments such as restaurants, improving the efficiency of outbreak investigations is critical to reducing illness and to developing better prevention strategies [Reference Naimi6, Reference O'Brien8, Reference Rossi, Faustini and Perucci9].
This study used clinical profiles to evaluate outbreaks of unknown aetiology reported in the United States from 1982 to 1997. In particular, we examined the potential impact of clinical profiling on the determination of contributing factors for outbreaks in restaurants during this time period.
METHODS
State and local health departments report results of foodborne outbreak investigations to the national foodborne disease outbreak surveillance system using a standard form. All confirmed foodborne outbreaks reported to the Centers for Disease Control and Prevention (CDC) from 1982 to 1997 were reviewed as previously described [Reference Hall2]. Regardless of aetiology, outbreaks were included if they involved at least five cases and had complete information on median periods of incubation and illness duration, and percentage of cases experiencing vomiting and fever. Outbreaks with known aetiology, defined by standard criteria, were used to develop the epidemiological profiles (Table 1) [Reference Olson1].
Table 1. Interquartile ranges for outbreak characteristics associated with pathogen profiles caused by known agents

* Profiles were compiled from outbreaks of known aetiology as follows:
• Vomiting toxin: Staphylococcus aureus and Bacillus cereus.
• Diarrhoea toxin: Clostridium perfringens.
• Salmonella-like: Salmonella enterica, Shigella spp., Campylobacter spp.
• Norovirus-like: Norovirus.
• E. coli-like: all confirmed E. coli outbreaks, regardless of serotype or virulence pattern.
Development and evaluation of clinical profiles
Clinical profiles were developed using a two-stage approach, modified from the methods used by Hall and colleagues [Reference Hall2]. As in Hall's study, the profiles were based on the following characteristics for each outbreak: median incubation, median illness duration, percentage of cases with vomiting, percentage of cases with fever, and the ratio of the percentages of cases with vomiting and fever (Fig.). However, exclusive clinical profile patterns were initially established to capture the defining features of the outbreaks (Table 2). These were based on CDC guidelines for confirmation of foodborne-disease outbreaks, and modified to provide for exclusive categories [Reference Olson1]. For example, outbreaks caused by staphylococcal enterotoxin typically feature very short incubation periods and a high proportion of cases with vomiting. Thus, an outbreak with a median incubation period within 2–5 h with >50% of cases vomiting, and a ratio of vomiting to fever >1·5 was assigned to the vomiting-toxin pattern. Outbreaks which did not exactly match one of these defined patterns were matched to one of five clinical profiles using a best-fit algorithm [Reference Hall2]. This was derived from interquartile ranges for the same clinical characteristics associated with known agents in the outbreak database (Table 1). For example, if an outbreak matched two characteristics of the vomiting-toxin profile, but no more than one characteristic of other profiles, it was assigned to the vomiting-toxin clinical profile.
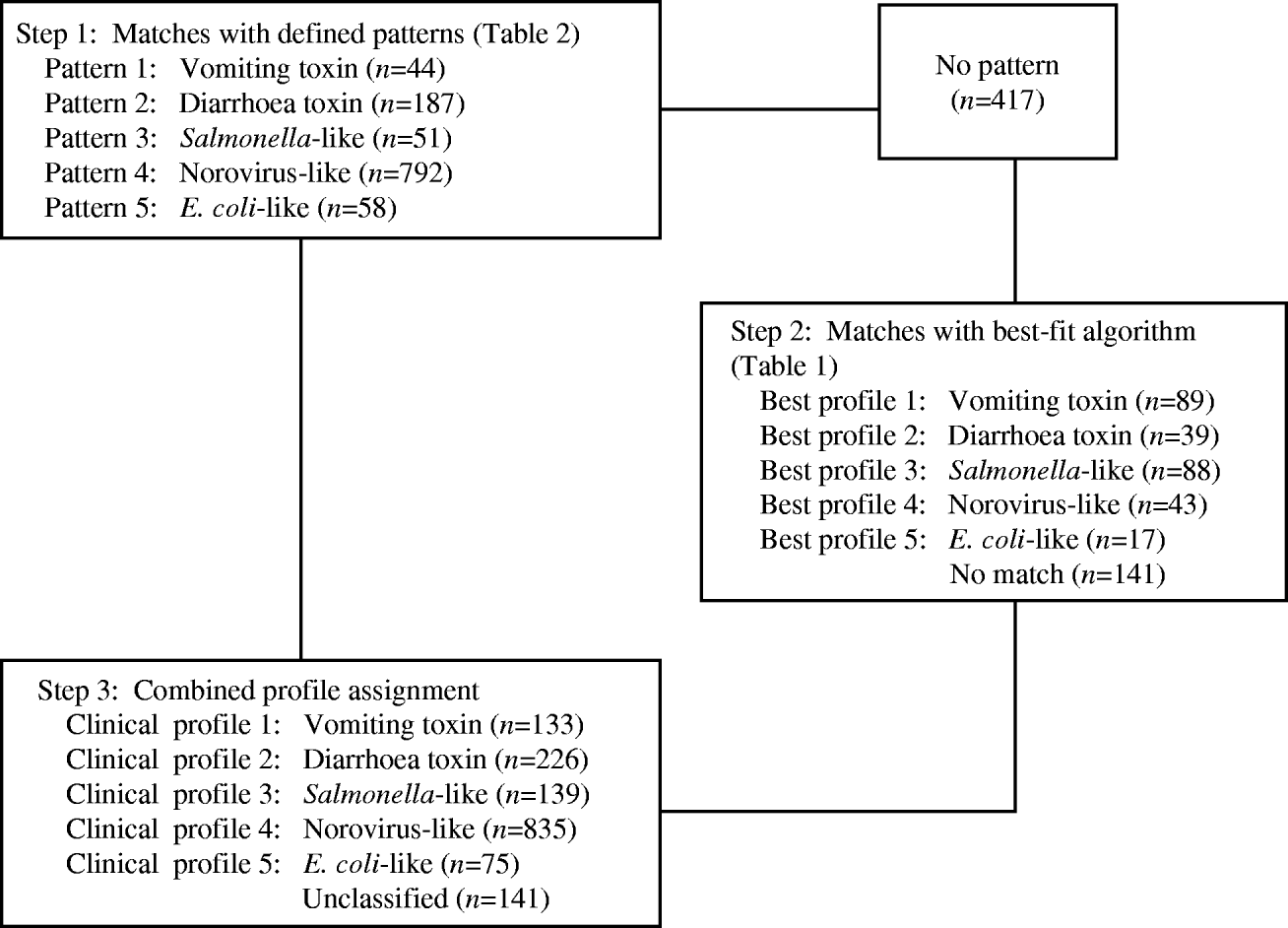
Fig. Clinical profile method of classifying outbreaks of undetermined aetiology into pathogen syndromes.
Table 2. Clinical criteria for classification of outbreaks into defined patterns

* Placement into the norovirus-like pattern required either a percent with vomiting of ⩾50%, or a percent with vomiting/percent with fever ratio of ⩾1.
Estimates of the sensitivity and specificity of the clinical profiles were determined by the ability of the profile to classify correctly outbreaks of known aetiology from which the clinical profile was derived.
Evaluation of contributing factors for outbreaks in restaurants
The outbreak database included information on five factors that have been identified as contributing to the occurrence of foodborne outbreaks [Reference Olson1]. These included: improper storage or handling temperature, inadequate cooking, contaminated equipment, poor personal hygiene of food handler, and food obtained from an unsafe source. These factors were recorded on standard outbreak reporting forms that were in use for national foodborne outbreak surveillance from 1973 to 1997 [Reference Olson1].
All outbreaks that occurred in restaurants were selected from the set of outbreaks used to develop and evaluate the clinical profiles. Outbreaks were excluded if information was missing for all five contributing factors. The percentage of outbreaks, by known agent and clinical profile, was calculated as the number of outbreaks in which the contributing factor was identified divided by the number of outbreaks evaluated.
To evaluate the association between known agents and contributing factors, we compared the rate of occurrence of each contributing factor in outbreaks in which aetiology was determined to their rate of occurrence in outbreaks that remained unclassified. Similarly, we compared the occurrence of contributing factors in outbreaks in which aetiology was assigned by clinical profile to their occurrence in the unclassified outbreaks. χ2 tests were performed to evaluate these proportions (Epi-Info 2002; CDC, Atlanta, GA, USA).
RESULTS
Between 1982 and 1997, 8781 foodborne-disease outbreaks were reported and entered into the CDC foodborne-disease outbreak surveillance database. Of these, 2246 (26%) involved at least five cases and had sufficient clinical information reported to be included in the study. A specific aetiology was confirmed by laboratory testing in 697 (31%) outbreaks; 1549 (69%) were categorized as undetermined aetiology (Table 3). A total of 454 Salmonella outbreaks accounted for 20% of all outbreaks and 65% of outbreaks with a known aetiology.
Table 3. Distribution of outbreaks of known aetiology selected for inclusion in the study
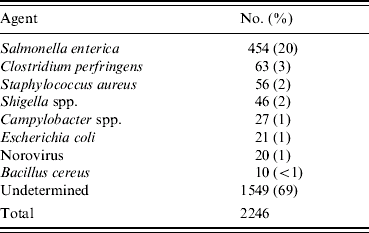
Clinical profiling of outbreaks
Of the 1549 undetermined aetiology outbreaks, 1132 (73%) were matched to a clinical profile based on one of the defined patterns. Sensitivities of the defined patterns in outbreaks of known aetiology ranged from 62% to 78%, with the lowest sensitivities found for vomiting-toxin outbreaks (Table 4). An additional 276 (18%) outbreaks were matched to a clinical profile using the best-fit algorithm. Overall sensitivities of the clinical profiles in outbreaks of known aetiology ranged from 70% to 87%, with the lowest sensitivity found for norovirus-like outbreaks (Table 4). Overall specificities ranged from 94% to 98%, with the lowest specificity found for E. coli-like outbreaks.
Table 4. Estimates of sensitivity and specificity for clinical profile classification of outbreaks with known aetiology
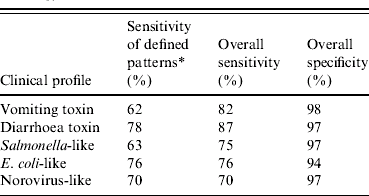
* Percentage of outbreaks with known aetiology classified into corresponding pattern based on criteria defined in Table 2. Overall sensitivity and specificity was based on the results of the combined two-step approach.
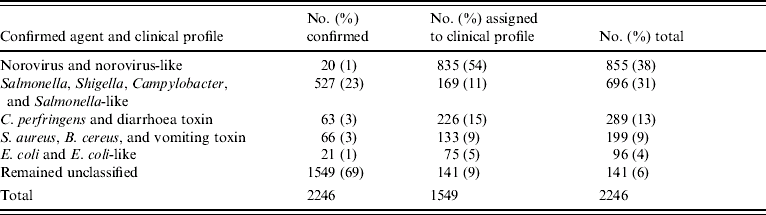
Among undetermined aetiology outbreaks, 835 (54%) fitted the norovirus-like epidemiological profile (Fig.). Thus, norovirus and norovirus-like outbreaks accounted for 38% of all outbreaks included in the study (Table 5).
Table 5. Distribution of outbreaks by known agent and clinical profile
Evaluation of contributing factors for outbreaks in restaurants
Of the 697 outbreaks with a laboratory-confirmed aetiology, 273 (39%) occurred in restaurants. Of the 1549 outbreaks with an undetermined aetiology, 678 (44%) occurred in restaurants. However, 10 (4%) of the restaurant outbreaks with known aetiology, and 77 (11%) of those with undetermined aetiology, had missing information for all contributing factors. Thus, 864 outbreaks in restaurants were used for this analysis (Table 6). Improper holding was cited as a contributing factor in 302 (35%) outbreaks, poor personal hygiene in 281 (33%), contaminated equipment in 167 (19%), and inadequate cooking in 156 (18%). Obtaining foods from an unsafe source was cited in only 6% of outbreaks and was dropped from the analysis.
Table 6. Contributing factors identified for outbreaks in a restaurant, excluding outbreaks with missing information for all contributing factors
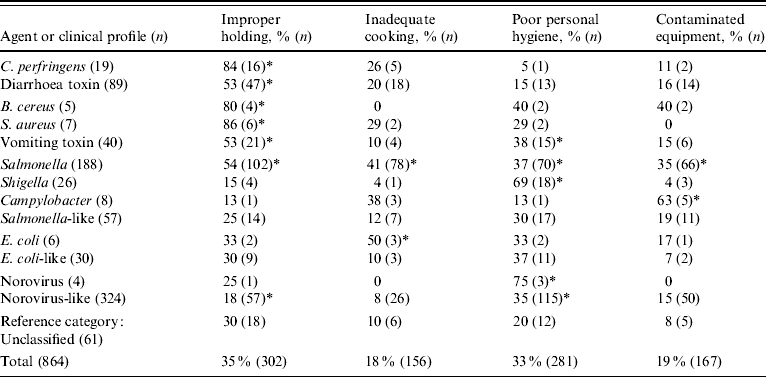
Percentage of outbreaks, by agent or clinical profile, with contributing factor identified, compared to percentage of unclassified outbreaks with contributing factor present: * P<0·05.
Contributing factors reported from outbreaks of known aetiology were consistent with the known biology of the agent (Table 6). Thus, improper holding time and temperature were cited as contributing factors for 84% of Clostridium perfringens outbreaks, but only for 15% of Shigella outbreaks. Similarly, poor personal hygiene was cited as a contributing factor in 69% of Shigella outbreaks, but in only 5% of C. perfringens outbreaks. Outbreaks of Salmonella were attributed to a broad range of contributing factors.
Similar associations were observed in the outbreaks of undetermined aetiology that matched a specific clinical profile (Table 6). Compared to outbreaks of undetermined aetiology that did not match a clinical profile, improper holding times and temperatures were significantly more likely to be identified in outbreaks caused by C. perfringens, B. cereus, S. aureus, and Salmonella (Table 6). Improper holding was also identified as a contributing factor more frequently in outbreaks with diarrhea-toxin and vomiting-toxin clinical profiles. In contrast, improper holding was less likely to be identified as a contributing factor in outbreaks with a norovirus-like clinical profile. Poor personal hygiene was more frequently associated with outbreaks caused by norovirus, Shigella, and Salmonella, and in outbreaks with norovirus-like and vomiting-toxin clinical profiles. Whenever the same contributing factor was associated with outbreaks caused by a known agent and with those corresponding to the agent's clinical profile, the contributing factor was identified in a higher percentage of outbreaks caused by the known agent than in those with the corresponding clinical profile.
DISCUSSION
This study refines and extends previous work in demonstrating that most outbreaks classified as having an undetermined aetiology have clinical characteristics resembling those of outbreaks caused by known agents [Reference Hall2]. In particular, a high proportion of these outbreaks appeared to be consistent with norovirus during the time of the study. The specificity estimates for the profiles ranged from 94% to 98%, suggesting a high degree of confidence in the assignment of a profile to an outbreak. The lower range of sensitivity estimates for the profiles suggest that many of the outbreaks that remained unclassified were probably also associated with known agents. In particular, the norovirus-like clinical profile accounted for 54% of the outbreaks originally classified as of undetermined aetiology, despite the fact that the norovirus-like profile had the lowest estimated sensitivity (70%). Although this estimate was based on a small number of outbreaks, the results are consistent with a recent evaluation of the role of norovirus in foodborne outbreaks reported to CDC from 1998 to 2000 [Reference Turcios10]. Turcios and colleagues found that clinical criteria for classifying outbreaks of norovirus had a specificity of 99% and a sensitivity of 68% [Reference Turcios10]. Thus, our estimate for the proportion of outbreaks attributable to norovirus is probably too low. In addition, recent improvements in the diagnostic capacity of state public health departments mean that a much greater proportion of norovirus outbreaks are now likely to be laboratory confirmed [Reference Turcios10]. Thus, it would not be appropriate to apply the 54% factor from 1982 to 1997 to the current outbreak situation.
Results of this study demonstrate the importance of determining the aetiological agent to assist in the environmental investigation of an outbreak in a restaurant. In practice, there is considerable variability in how EHS determine that a particular food-handling problem represents a contributing factor to the occurrence of a foodborne outbreak. In outbreaks where an agent is already known, the EHS may go into the investigation with predetermined knowledge about what the contributing factors are likely to be. Thus, they will be looking for, and as a result may be more likely to find, a contributing factor that corresponds to the aetiology. Our findings suggest that this is a common practice in outbreak investigations. For example, in 84% of outbreaks caused by C. perfringens, improper holding times and temperatures were identified as contributing factors. This finding is consistent with the fact that improper holding is necessary for sufficient growth of C. perfringens to reach explosively infectious levels on the contaminated food item. In contrast, Shigella has a lower infectious dose and humans are the only source of contamination. Thus, improper holding would not be expected to contribute to outbreaks of Shigella, and in fact it was identified in only 15% of Shigella outbreaks, whereas, poor personal hygiene was identified in 69% of them. Interestingly, improper holding, inadequate cooking, poor personal hygiene and contaminated equipment all appeared to be important for transmission of Salmonella. This finding is consistent with the multiple possible sources for contamination, environmental persistence, and relatively low infectious dose for Salmonella infection.
Our results suggest that EHS use information about the aetiology of an outbreak to guide their evaluations of restaurants. It has not previously been clear how EHS identify contributing factors in outbreaks when no agent is confirmed by laboratory testing. In investigations where the aetiology of the outbreak is truly unknown, the EHS may act as an impartial reviewer of the situation, without selectively focusing on any particular food-handling practices. In this type of investigation, the EHS will probably observe common food-handling problems and may observe the actual problems that led to the occurrence of the outbreak. However, without any knowledge of the aetiology, the EHS may not be able to distinguish one from the other and will either report all observations or none of them. For example, a baseline survey of retail food-handling practices conducted by the Food and Drug Administration found high rates of food-handling errors during routine evaluations [11]. Improper holding times and temperatures were noted in 63% of observations made in full service restaurants and poor personal hygiene practices were noted in 53%. Without knowledge of a specific agent, such observations could be assumed to be contributing to the occurrence of an outbreak. In fact, these were also the most common contributing factors cited in our study for outbreaks of undetermined aetiology that did not match an epidemiological profile. However, improper holding times and temperatures were cited in only 30% and poor personal hygiene in only 20% of these unclassified outbreaks. This suggests reluctance by EHS to identify contributing factors when the relationship between the observed food-handling problem and the aetiology cannot be determined.
Furthermore, in outbreaks of unknown aetiology in which the clinical information suggests an aetiology, our results demonstrate that EHS empirically use this information to guide their evaluations. Thus, outbreaks with diarrhea-toxin and vomiting-toxin profiles were more likely to have improper holding identified as a contributing factor than were outbreaks that remained unclassified. Furthermore, outbreaks with a norovirus-like clinical profile were less likely to have improper holding identified as a contributing factor. In the same manner, outbreaks with a norovirus-like clinical profile were more likely to have poor personal hygiene identified as a contributing factor than were the unclassified outbreaks. These data demonstrate that biologically plausible factors were attributed based on probable aetiology in the absence of laboratory evidence. In contrast, outbreaks matching Salmonella-like or E. coli-like clinical profiles were similar to the unclassified outbreaks with respect to contributing factors identified, consistent with the fact that the biology of these organisms does not lead to a strong a priori hypothesis about the probable source.
These findings have three important implications for outbreak investigations and surveillance. First, the use of clinical profiles is important in guiding environmental evaluations of establishments involved in outbreaks. Identifying a probable aetiology can help narrow the focus of the investigation to better identify the source and the environmental antecedents leading to its occurrence. This is a critical step in improving prevention measures. While unbiased observations of food-handling errors in an outbreak setting may identify many targets for remediation, the failure to focus on the specific problem that led to the occurrence of the outbreak may leave the restaurant at greater risk for recurrence of a similar event. The EHS-Net, a collaborative network of EHS from CDC and nine states, in association with FoodNet, conducted outbreak and non-outbreak environmental evaluations in restaurants to provide a systematic framework for understanding these events. Although pathogen-specific contributing factors were identified, outbreak and non-outbreak restaurants were similar with respect to many characteristics [Reference Hedberg12]. Second, by identifying agents for which specialized laboratory testing may be needed, the use of clinical profiles may help guide diagnostic testing and thus enable laboratory confirmation of the agent. This can also potentially lead to the identification of new and emerging pathogens. Third, and most importantly, clinical profiles of outbreaks can usually be obtained by interviewing cases within hours of initial notification. Thus, this information can be rapidly used to guide investigations, often well before laboratory results are available. Given the importance of rapidly and effectively responding to outbreaks of foodborne disease, this is a major consideration.
While results of this study suggest that EHS are using clinical profiles to guide outbreak investigations in restaurants, it is also clear that relevant contributing factors are less likely to be identified in these outbreaks than in the corresponding outbreaks for which an agent is confirmed by laboratory testing. This suggests either that the use of clinical profiles is not as widespread as it could be, or that there is less confidence in reporting results based on these findings. More widespread use of simple clinical profiles, such as those defined in Table 2, may promote the timeliness and efficiency of outbreak investigations. Presenting data based on clinical profiles in surveillance summaries will provide state and local agencies with an incentive to incorporate these methods and report their results.
Improving the basis for relating contributing factors to suspected aetiology should also provide an incentive to improve reporting of contributing factor data. In 1998, the national foodborne disease surveillance reporting form, containing five contributing factors, was replaced with a much more detailed form including 14 factors that related to contamination of foods, five factors associated with survival or lack of inactivation, and 12 factors associated with proliferation and amplification [Reference Bryan, Guzewich and Todd13]. Improved ascertainment of aetiology and contributing should strengthen the public health system's contribution to safeguarding the food supply.
ACKNOWLEDGEMENTS
This work was supported by a cooperative agreement between the CDC and the Association for Schools of Public Health, Project No. S1855. The authors thank Dr Christopher Braden, and Alana Sulka, CDC, Foodborne Outbreaks Response and Surveillance Unit for assistance in obtaining and analysing the outbreak surveillance data.
DECLARATION OF INTEREST
None.











