INTRODUCTION
Penguins are long-lived aquatic birds exclusively distributed in the Southern hemisphere and are catalogued as marine sentinels of the ocean's health. This condition has been established, among other reasons, due to their large land breeding colonies [Reference Boersma1], and being totally dependent on marine resources [Reference Trivelpiece2]. Therefore their population alterations reflect the regional oceanic variations more accurately and faster than any other aquatic bird [Reference Boersma1].
The Spheniscus genus includes four species which inhabit the coastal areas from the Pacific and Atlantic Oceans. The Magellanic penguin (Spheniscus magellanicus) is distributed in southern South America, including Chile and Argentina. It is the most abundant temperate penguin in the world [Reference Boersma1], although with a declining population that has caused its ‘Near Threatened’ classification by the International Union for the Conservation of Nature [3]. During the reproductive season (spring and summer) it is possible to observe colonies from 30° S in the Pacific and 42° S in the Atlantic coasts. In winter, birds migrate to Brazil, South Atlantic Islands, New Zealand and Australia [Reference Miranda4].
It has been assumed that emerging infectious diseases (EIDs) constitute an unpredictable phenomenon associated with global changes largely influenced by anthropogenic effects on the environment, hosts and pathogens [Reference Rhyan and Spraker5]. In this scenario, penguins are seriously exposed due to resource competition with commercial fisheries and the progressive human invasion of their habitats through population growth and development, including livestock, mining and touristic activities. In addition, the contamination of rivers and oceans with sewage and fishery processing waste predisposes contact with biological agents, threatening the sanitary condition and conservation status of penguins [Reference Boersma1]. This situation is particularly important for temperate penguins like S. magellanicus in Chilean Patagonia, which are a tourist attraction, increasing the direct and indirect interaction with visitors, domestic animals and their transmissible pathogens.
Worldwide, the need for active surveillance of wildlife-borne pathogens has been recognized, focusing these efforts on prioritized agents. Salmonella enterica has been considered within this group, being associated with both water and wildlife [Reference Humblet6, Reference Tavernier7]. Within the World Health Organization (WHO) Event Management System, salmonellosis is classified as a communicable disease common to humans and animals related to food safety, with transmission through the food chain and water supply [Reference Schneider8]. In developed and developing countries, Salmonella constitutes an endemic foodborne infection that generates periodic outbreaks in the human population and is considered of great concern for animal health [Reference Tavernier7, Reference Cardoen9, Reference Hendriksen10]. The most frequent Salmonella serovars isolated from wild birds (mainly aquatic birds) are S. enterica Typhimurium and S. enterica Enteritidis [Reference Hubalek11–Reference Pennycott, Park and Mather13]. Although in most cases the infection is asymptomatic, these serovars have been associated with disease outbreaks and high mortality [Reference Hubalek11, Reference Hall and Saito14–Reference Reed16], with reports evidencing a direct transmission of Salmonella from wild birds to humans and other animals [Reference Dhama, Mahendran and Tomar17–Reference Reche19].
The aim of this work was to detect S. enterica serovars in faecal samples of free-ranging Magellanic penguins from two colonies in Chilean Patagonia. The isolates were characterized by serotyping, antimicrobial susceptibility and genotyping.
MATERIAL AND METHODS
Study area
During January and February in 2012 and 2013, samples from S. magellanicus located in Magdalena Island (52° 55′ S, 70° 34′ W) and Otway Sound (52° 58′ S, 71° 13′ W), in southern Chilean Patagonia were collected. For this work, authorization for sampling activities were obtained from the official authority in Magdalena Island and from farm owners in Otway Sound. Additionally, authorizations from Bioethics and Biosecurity Committees of the University of Chile were also obtained.
Sampling
A total of 2114 faecal samples (Table 1) were collected through both environmental and cloacal swabbing. Swabs were placed into Cary–Blair transport medium (Copan, USA) and stored under refrigeration for up to 4 weeks until arrival at the Laboratory of Infectious Diseases, University of Chile, Santiago.
Table 1. Salmonella strains isolated from Magellanic penguins (Spheniscus magellanicus)

AMP, Ampicillin; CTX, cefotaxime; EFT, ceftiofur; TE, tetracycline.
Penguins were captured using a net, following recommendations of Chilean authorities and the Global Penguin Society. In order to avoid breeding interference, only adult animals ranging outside nests were manipulated. Once cloacal swabbing and morphological data collection was completed, the animals were identified by a web tag (National Band and Tag Co. model no. 1005–1) as described previously [Reference Boersma and Rebstock20], and then released in the same place as captured.
Bacterial isolates
To isolate bacteria, swabs were placed into 5 ml buffered peptone water (Difco APT broth, Beckton Dickinson, USA) supplemented with 20 μg/ml novobiocin (Sigma, USA) [ Reference Jensen 21 ], and incubated for 24 h at 37 °C. Then, 100 μl of the suspension was inoculated into modified semi-solid Rappaport–Vassiliadis basal medium (Oxoid, Brazil) supplemented with 20 μg/ml novobiocin and incubated for 24 h or 48 h at 41·5 °C. Cultures with growth were plated onto xylose lysine deoxycholate agar (Difco XLD, Beckton Dickinson) and incubated for 24 h at 37 °C. Suspicious colonies were identified by biochemichal tests and invA gene detection by PCR [Reference Malorny22]. Next, S. enterica strains were serotyped according to the Kauffman–White scheme [Reference Grimont23].
For genotypic comparison, a set of S. enterica strains isolated from poultry and humans in Chile during 2011 and 2012 were also included in the analysis (Fig. 1). Strains from poultry were provided by the Agriculture and Livestock Service (SAG) and strains from humans by the Institute of Public Health (ISP). A S. enterica Agona strain previously isolated from a Kelp gull (Larus dominicanus) [Reference Fresno24] was also included.
Genotyping
Pulsed field gel electrophoresis (PFGE)
PFGE was performed according to the standard protocol recommended by PulseNet (http://www.cdc.gov/pulsenet/pathogens/index.html). Briefly, the digestion was made using XbaI (Invitrogen, USA). Electrophoresis was performed using the CHEF DRIII PFGE system (Bio-Rad, USA). The conditions used were 6 V/cm for 21 h at 14 °C with pulse time ranging from 3 to 63 s. As control, S. Braenderup H9812 strain was used. The gels were analysed with GEL COMPAR II® software (Applied Maths, Belgium).
Virulotyping
This procedure was made by PCR amplification of pefA, spvC, sirA, gipA, SEN1417, trhH and prot6e virulence genes, using primers pefA_F (5′-cctgtgacctgaccacttctg-3′), pefA_R (5′-gtaagccactgcgaaagatg-3′), spvC_F (5′-ctccttgcacaaccaaatgcg-3′), spvC_R (5′-tgtctctgcatttcaccaccatc-3′), sirA_F (5′-tgcgcctggtgacaaaactg-3′), sirA_R (5′-actgacttcccaggctacagca-3′), gipA_F (5′-acgactgagcaggctgag-3′), gipA_R (5′-ttggaaatggtgacggtagac-3′), sen1417_F (5′-gatcgctggctggtc-3′), sen1417_R (5′-ctgaccgtaatggcga-3′), trhH_F(5′-aactggtgccgttgtcattg-3′), trhH_R (5′-gatggtctgtgcttgctgag-3′), prot6e_F (5′-gcctaaggttagtgtgactctc-3′) and prot6e_R (5′-ctagcagccgttggtatcc-3′). The DNA extraction, reaction mixtures and PCR conditions were developed as described previously [Reference Fresno24]. The S. enterica Typhimurium ATCC 14028 and S. enterica Agona SARB1 strains were used as positive controls.
Antimicrobial resistance phenotypes
Antimicrobial susceptibility was evaluated by the disc diffusion method following CLSI criteria [25]. Antimicrobials tested were (μg/disk) ampicillin (10), amoxicillin–clavulanic acid (20/10), cefotaxime (30), gentamicin (10), trimethoprim–sulfamethoxazole (1·25/23·75), tetracycline (30), ciprofloxacin (5), cefradine (30), ceftiofur (30) and enrofloxacin (10) (Oxoid). Escherichia coli ATCC 25922 was used as control strain.
Statistical analyses
Categorical data analyses were made through contingency tables with Infostat (2010v) software (http://www.infostat.com.ar/) using Pearson's correlation coefficient to determine differences (P < 0·05).
Results from PFGE and PCR were merged by transforming data in a binary code, using 1 when the character was present (PFGE fragment or PCR gene detection) or 0 when it was absent. The similarity of the strains was calculated according to the Dice coefficient with a 1% tolerance in band position, and the dendogram was constructed using the UPGMA method with TREECON software [Reference Van de Peer and De Wachter26].
RESULTS
Three S. enterica Agona and five S. enterica Enteritidis strains were isolated, with an overall infection rate of 0·38%. The detection of these serotypes suggest differences according to sampling region and year (Table 1), because Enteritidis strains were only detected in Otway Sound, and Agona strains only in 2012. In addition, phenotypes of resistance against ampicillin, cefotaxime, ceftiofur and tetracycline were detected (Table 1). Resistances to ceftiofur and tetracycline were associated (P < 0·05) with Agona and Enteritidis strains, respectively.
The PFGE assay classified the strains in two main clusters, in accordance with their serotypes, with less than 60% similarity between them (Fig. 1). When the PFGE and PCR results were merged (Fig. 2), these major groups were maintained and discrimination improved marginally, with some strains changing from identical patterns to having minor differences (less than 10%) within sub-clusters. Within the two sub-clusters of the S. enterica Enteritidis group, all penguin isolates were distinguishable from strains belonging to other hosts. However, similarities were ⩾95%, suggesting genetically close Salmonella strains. Within the S. enterica Agona group, strains from humans were clearly differentiated from the others, and one penguin's isolate (S. enterica Agona 6) had an identical pattern with a gull's strain (S. enterica Agona 2). When comparing serotypes, a higher genotypic variability in S. enterica Agona strains was found (Figs 1 and 2) compared to the S. enterica Enteritidis group.
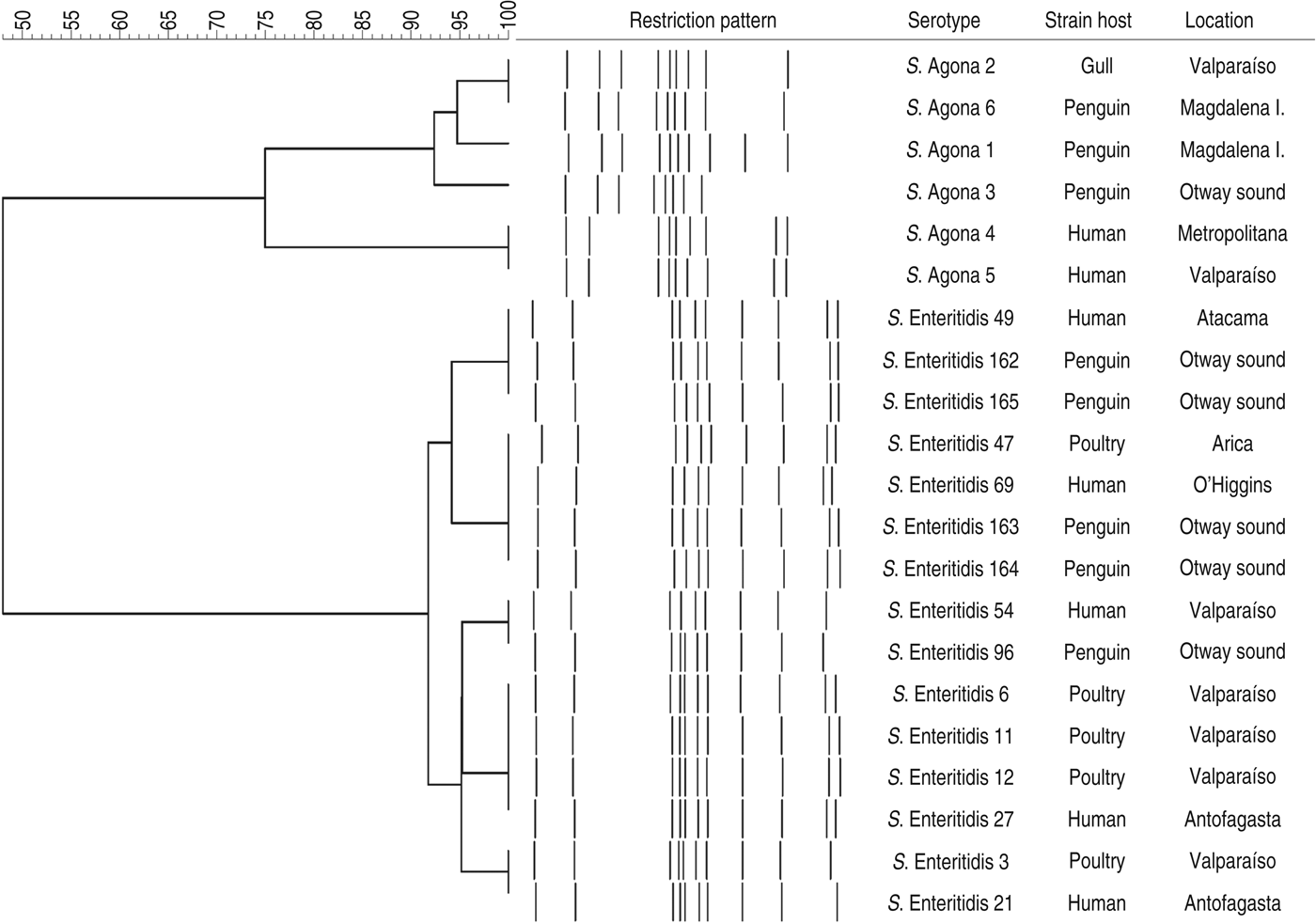
Fig. 1. Dendogram showing genetic similarities (%) between Salmonella enterica strains resulting from PFGE assay after digestion with XbaI. The tree was constructed using the Dice coefficient and UPGMA algorithm with Gel Compar software (Applied Maths, Belgium).
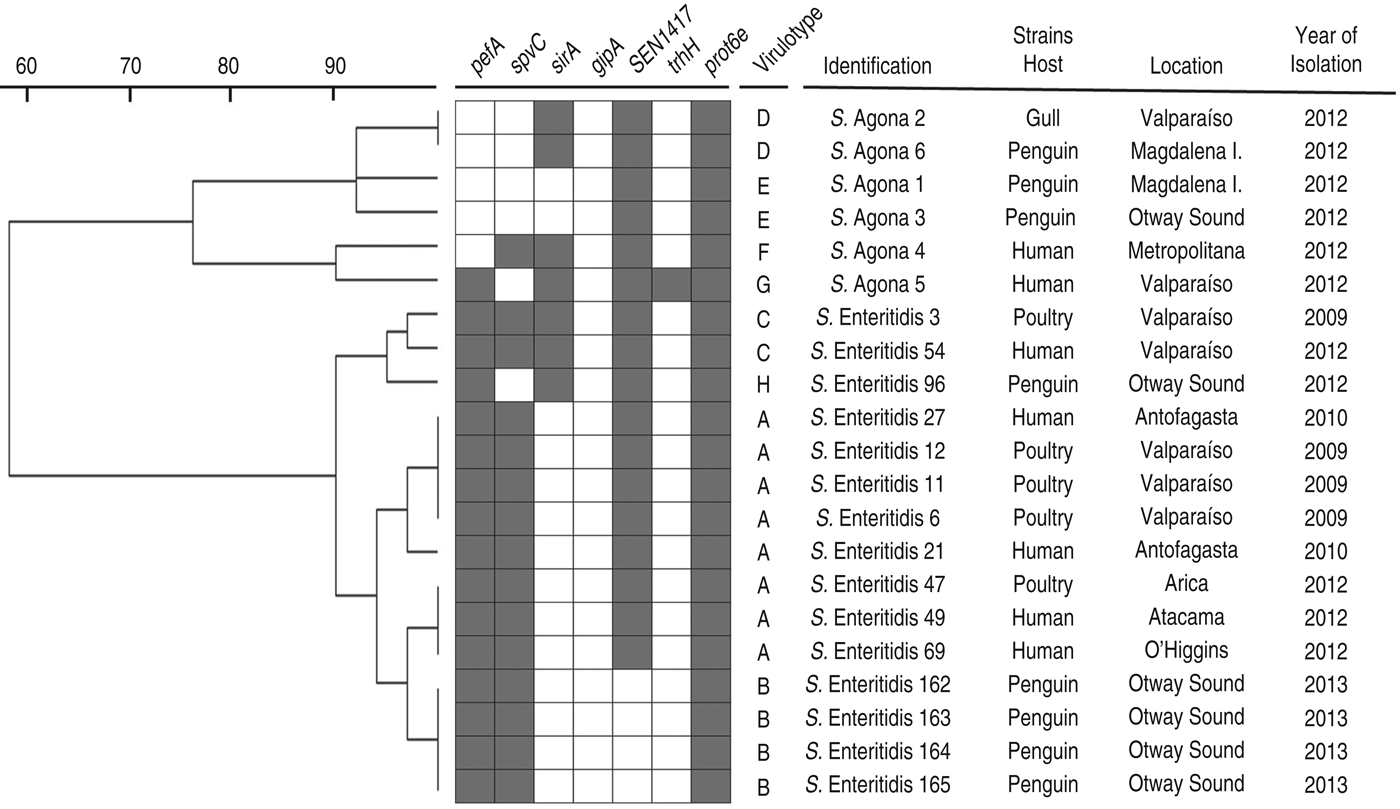
Fig. 2. Dendogram showing genetic similarities (%) between Salmonella enterica strains resulting from merged PFGE and PCR data. Detection of virulence-associated genes is depicted as grey squares when present. Virulotypes are indicated by letters according to their frequencies (A:8, B:4, C to E:2, F to H:1). Results from PFGE and PCR were transformed into a binary code, using 1 when the character was present (PFGE fragment or PCR gene detection) and 0 when absent. The tree was constructed using the UPGMA method with TREECON software.
DISCUSSION
It is now estimated that 70–80% of emerging infectious diseases (EID) in humans have an animal component in their transmission, and more than half of these have been elicited from wildlife [Reference Jones27]. Moreover, the most common cause of EID in wildlife is the human introduction of pathogens in wild environments [Reference Gilbert, Slingenbergh and Xiao28, Reference Greger29], among which S. enterica is within the group of pathogens that deserve more attention for surveillance [Reference Tavernier7].
The presence of Salmonella in penguins has been reported in Pygoscelis adeliae from the Antarctic continent, including serotypes Blockley, Panama and Infantis with an overall infection rate of 13%. Other studies have reported Salmonella infection in P. papua, with serotypes Enteritidis, Havana and Typhimurium, with isolation rates ranging between 3% and 44% [Reference Dimitrov, Metcheva and Kenarova30, Reference Olsen31]. These works, with an overall 108 positive samples, suggest that P. papua could be a major reservoir of the bacterium in Antarctica, contrasting with penguin species Aptenodytes forsteri and Eudyptes chrysolophus that were also sampled without any detection [Reference Palmgren32, Reference Oelke and Steiniger33]. In this study we detected a very low occurrence of Salmonella infection in S. magellanicus (0·38%), with technical, geographical and host-related factors that could explain this contrasting result. Nonetheless, reports made from one penguin species cannot be extrapolated to others, which is also supported by a recent description of large variation within the faecal microbiota of different penguin species, a finding that includes bacteria belonging to the family Enterobacteriaceae and several known human pathogens [Reference Dewar34].
This is the first report of Salmonella infection in free-ranging penguins from South America, and the first description of antimicrobial resistance phenotypes among these isolates. However, a previous work has already shown tetracycline and ampicillin antibiotic resistance in enteric non-pathogenic microorganisms isolated from penguin faecal samples collected near human settlements in Antarctica [Reference Miller, Gammon and Day35]. This finding, and the fact that tetracycline and ampicillin correspond to widely used antimicrobials in humans, livestock and poultry [Reference Benacer36], suggest that these bacterial phenotypes in penguins, which inhabit pristine environments, could be a measure of the anthropogenic effect. However, whether penguins are directly or indirectly being affected by these human footprints is unknown, since other seabirds in Chile have also been reported to harbour Salmonella strains with these and other antimicrobial resistance phenotypes [Reference Fresno24]. This hypothetical inter-species transmission gains support with our genotypic results, in which one S. enterica Agona strain isolated from a Kelp gull on the Chilean coast, had the same pattern as a penguin isolate (Figs 1 and 2). Furthermore, all S. enterica Enteritidis strains isolated from penguins showed high genetic similarity with other human and poultry isolates (Fig. 2), representing additional evidences of close bacterial transmission cycles among hosts.
The analysis based on merged PFGE and PCR results has clearly discriminated bacteria according to their serotypes. The contrasting strains' diversity at the sub-cluster level is in agreement with the relative higher clonality that has been reported for S. enterica Enteritidis strains compared to other serotypes [Reference Thong37, Reference Zheng38].
On the other hand, the SEN1417 factor has been identified as a putative ABC transporter protein, which gene is inserted within an unstable chromosomal segment that has been exclusively detected in prevalent phage types isolated from humans and animals [Reference Pan39]. In this study, this sequence was detected in all S. enterica strains isolated during 2012 and none from 2013. Whether this is by coincidence or bacterial evolution remains unknown and should be elucidated in the future.
In Chilean Patagonia, there are wild colonies of S. magellanicus neighbouring urban zones with high tourist activity, which increases in the summer during the reproductive period of the animals. It has been calculated that almost 3 00 000 people per year, from all over the world, visit the studied region. However, other activities such as mining, animal husbandry and maritime traffic have been developed near penguins' habitats. The fact that reported Enteritidis and Agona serotypes are among the top five most common zoonotic serotypes isolated from humans in the South American region [Reference Hendriksen10], along with the antimicrobial resistance phenotypes (Table 1) and genotypic similarities detected (Figs 1 and 2), constitute suggestive evidence that penguins have been exposed to human influence in Patagonia. Further efforts are required to characterize the real magnitude of this phenomenon, the temporal and geographical fluctuations of Salmonella in penguin colonies, the sanitary effects on these animals, the involvement of other wildlife and the potential for the emergence of new Salmonella strains. This knowledge will support decisions to preserve the environment and its wildlife in a globally changing scenario.
DECLARATION OF INTEREST
None.
ACKNOWLEDGEMENTS
We thank Claudia Godoy, Rigofredo Veneros and Marcela Fresno for their help during field and laboratory work. We also thank Mary Kalb, for critical review of this paper, and Alda Fernández (Instituto de Salud Pública) for providing Salmonella strains isolated from humans. This work was supported by Fondecyt project 1 110 255 and predoctoral fellowships from CONICYT.








