INTRODUCTION
Escherichia coli O157 infection is a significant cause of human illness in Canada, particularly in the province of Alberta. In 2004, the overall rate of E. coli O157:H7 infection for Canada was 3·36/100 000 population, while Alberta's rate was more than double at 8·96/100 000 population [1]. E. coli is a species of bacteria which is naturally present in human and animal faeces. However, some strains, such as E. coli O157:H7, produce a special type of toxin that can cause severe illness in humans [Reference Mead and Griffin2, Reference Tarr3]. Cattle serve as a recognized reservoir for E. coli O157, as the organism can reside asymptomatically in the animal's intestinal tract [Reference Besser4]. The principal source of human infection is due to faecal contamination of food and water sources. Incorrectly prepared hamburger has been identified as a common source of food-associated infection [Reference Mead5, Reference Waters, Sharp and Dev6]. Outcomes of infection include bloody diarrhoea and haemolytic uraemic syndrome (HUS), a significant cause of acute renal failure in children [Reference Chan, Williams and Roth7]. Perhaps the most well-known Canadian outbreak of E. coli O157 resulted from the contamination of the water supply of Walkerton, Ontario in 2000. At least seven deaths were directly attributable to drinking contaminated water; thousands of people fell ill, and some are still suffering as a result [Reference Auld, MacIver and Klaassen8].
Several studies have shown a significant spatial association between cattle density and incidence of human E. coli O157 infection in Ontario [Reference Michel9, Reference Valcour10] and eastern Canada [Reference Valcour11]. Human E. coli O157 infection appears to have a similar geographical pattern to that of cattle density in Ontario [Reference Michel9]. Additional likely sources of infection are direct contact with infected cattle, or consumption of contaminated drinking water. The latter can be particularly problematical in instances of high rainfall, which can increase opportunities for water run-off and subsequent drinking-water contamination [Reference Jokinen12, Reference Thomas13].
Forty per cent (5·41 million head) of the Canadian cattle population currently reside in Alberta [14]. Few published studies of the relationship between human E. coli O157 infection and cattle density in Alberta are available in the scientific literature. Work by Pearl et al. [Reference Pearl15] presented inconclusive results regarding the impact of cattle density on human E. coli O157 in Alberta. Statistical clusters of higher disease rates were found covering large feedlots in the southern counties of Alberta; however, the clusters differed depending on the type of cases used in the analysis [Reference Pearl15]. Similarly, a search of the current literature reveals no published studies of the relationship between human E. coli O157 infection and weather events in Alberta. Research by Jokinen et al. [Reference Jokinen12] on bacterial pathogen contamination of surface waters in the Oldman River basin in southern Alberta demonstrated that E. coli O157:H7 prevalence in surface water was best characterized by the total rainfall on the day of sampling and 3 days in advance of sampling. However, relationships between extreme precipitation and human E. coli O157 in Alberta have not been directly studied. Curriero et al. [Reference Curriero16] studied the relationship of waterborne disease outbreaks with extreme rainfall in the USA and showed that 68% of the waterborne disease outbreaks, including E. coli O157, were preceded by extreme precipitation events above the 80th percentile. Understanding the associations and interactions between human E. coli O157 infection, livestock and weather events will help enable targeted preventative measures.
The primary objective of this study is to investigate the extent to which proximity to cattle and weather events in Alberta predispose human populations to E. coli O157 disease. This objective will be addressed by examining the relationship of livestock and weather variables with that of human E. coli O157 incidence rates of reported cases in Alberta between 2004 and 2011. The relationship between livestock, weather variables and disease data will be evaluated via regression models with varying methodologies for incorporating spatial information.
METHODS
Data sources
Cases of E. coli O157 between 2004 and 2011 in Alberta were obtained from the province's Communicable Disease Reporting System (CDRS). Each case consists of a Notifiable Disease Report that is completed by local public health departments and submitted to Alberta Health and Wellness. In addition, the Alberta Provincial Laboratory for Public Health conducts enteric pathogen screening and serological tests on samples obtained from these cases. Cases in CDRS therefore consist of individuals testing positive for E. coli O157 or presenting clinical signs and have been epidemiologically linked to a CDRS pre-existing case. Additional information regarding the presentation-diagnosis-reporting pyramid for E. coli verotoxigenic infection can be found on the Alberta Health and Wellness website [17].
Data obtained on each CDRS case consisted of sex, categorical age, symptom onset date, census consolidated subdivision (CCS) location, outbreak association and travel history. Categorical age was defined by the following groups: 0–9, 10–19, 20–29, 30–59, and ⩾60 years. Individuals suspected of becoming infected while outside Alberta, not residing in Alberta, or associated with a person-to-person outbreak were excluded from the analysis. These individuals were removed from the data as only environmental sources of infection in Alberta were of interest. An outbreak was defined as any grouping of more than three cases that were epidemiologically linked by Alberta Health and Wellness [17]. Only one such outbreak existed in the CDRS data. In this outbreak the first case according to symptom onset date was retained and the eight other linked cases were removed. The final dataset contained 1205 cases of E. coli O157 meeting the criteria described above.
Hospital inpatient records from the Discharge Abstract Database (DAD) were also obtained from Alberta Health and Wellness for cases presenting International Classification of Disease (ICD-10) codes (see Table 1), in any diagnosis field. ICD-10 codes not specific for E. coli O157 were selected to obtain cases for which E. coli O157 infection resulted in symptoms that were reported as non-specific. However, these ICD-10 codes also allow for cases not related to E. coli O157 infection to be included in the data. As such, selection of the ICD-10 codes was intended to reflect a balance between false-positive and false-negative results. A similar set of ICD-10 codes has been used previously [Reference Parmley18]. Cases obtained serve as a sensitive E. coli O157 dataset, hereafter referred to as the DAD dataset. Caution must be exercised when interpreting and utilizing the DAD as the true nature of each case is unknown and thus subject to bias. Data obtained on each case included sex, categorical age, hospital admittance date, CCS location, and Alberta residency status. Individuals defined as not being residents of Alberta were removed from the study. The DAD contains 4161 cases having experienced a disease listed in the ICD-10 codes listed in Table 1. Thirty cases of HUS were present in the DAD. The ethics board at the University of Guelph approved the present study and all data obtainment.
Table 1. International Classification of Disease (ICD-10) codes obtained from the discharge abstract database

Demographic data enumerating the age and CCS distribution of the Alberta population was obtained from the 2001, 2006 and 2011 Canadian Censuses of Population [19–21]. Age and CCS distribution were estimated for each study year by fitting a linear model between the encompassing census year's data and interpolating the results. A CCS consists of a group of adjacent census subdivisions (CSD) or if large enough geographically (>25 km2) or if a sizable population (>100 000) will consist of only one CSD. More than one CCS will combine to form a census division (CD). In Alberta there are 77 CCS and 19 CD. CCS type was determined based on the number of municipalities adjacent to an urban core having a population of 10 000–99 999 or >100 000. This is a modified version of Statistics Canada's statistical classification type [22]. A CCS type of census agglomeration has at least one municipality adjacent to an urban core with a population between 10 000 and 99 999 but no municipalities with a population ⩾100 000. The census metropolitan area (CMA) contains at least one municipality adjacent to an urban core with a population >100 000. CD type was created in the same fashion as the CCS type. A map depicting the location of Alberta's CCS and CD is available from Statistics Canada (http://www.statcan.gc.ca/ca-ra2006/m/alberta2-eng.pdf).
Livestock data were obtained from the 2001, 2006 and 2011 Canadian Censuses of Agriculture [23–25]. Like the demographic data, each study year's livestock data were estimated by fitting a linear model between the encompassing census year's data and interpolating the results. Table 2 presents a list of all cattle variables considered for inclusion in the final models. Each cattle variable was reported in three formats to correspond with the various analyses being conducted: the average number of cattle by CD, the average number of cattle by CCS and the monthly number of cattle by CD, where each month in a given year is equal. Livestock variables other than cattle were also obtained. These livestock variables included chicken, turkey, other poultry, pig, sheep, horse, goat, bison, deer, elk and llama density. Each animal density measure consisted of the total number of an animal in a given CD divided by the area (km2) of the CD, and were reported in formats of yearly density and average density. Other poultry consisted of geese, ducks, roosters, ostriches, emus, pheasants and quail. Livestock data, other than cattle, were only analysed at the CD level as the CCS level contained excessive missing values. Statistics Canada's policies on protecting the confidentiality of the livestock data led to the amalgamation of some adjacent CD and CCS. Thus the present study contains 69 CCS and 18 CD.
Table 2. Glossary of cattle variables used in the present study

Monthly weather data were obtained for all of the CD in Alberta. Data were collected from Canada's online National Climate Data and Information Archive [26]. Weather stations were selected so that they approximated the middle of the CD. Weather variables included monthly cumulative rainfall (mm), monthly cumulative precipitation (mm), mean maximum temperature (°C), mean temperature (°C) and an extreme rain days count. A mean maximum temperature is defined as the average of all daily maximum temperatures in a given month. The extreme rain days count was defined as the number of days in a given month that received >10 mm of rain. These weather variables were reported per month and CD as well as an overall average per CD for the full study period. A season variable was also created with spring (March–May), summer (June–August), autumn (September–November), and winter (December–February).
Statistical analyses
Statistical analysis was based on regression models. Various regression models were fitted in order to determine the best-fitting models. Linear regression models were constructed using three methods: ordinary least square (OLS) procedure with White's adjustment of the variance matrix, maximum-likelihood estimation (MLE) including a spatial lag coefficient and MLE including a spatial error coefficient. Each of these models has its own methodology in accounting for the spatial structure, which violates the homoscedastic and uncorrelated error assumptions of linear regression models. Poisson and negative binomial regression models were also used to incorporate the count nature of the data. The OLS and count models incorporated linear effects of latitude and longitude coordinates for each division's centroid, thus helping control for the spatial aspects of the data. The first three regression techniques were used by Michel et al. [Reference Michel9] and the count techniques were applied by Haus-Cheymol et al. [Reference Haus-Cheymol27]. For further technical details, please refer to these two papers. Spatial weights used in the spatial lag and error models were based on queen continuity. Queen continuity defines a neighbour as having a common boundary and/or vertices with the region of interest [Reference Anselin28]. Spatial weights, and latitude and longitude of each division's centroid were obtained using OpenGeoDa [Reference Anselin29].
Monthly incidence rates were modelled using both zero-inflated Poisson and negative binomial models. These models attempt to account for excess zeros by utilizing two sub-models, one to account for the excess zeros and one to model the counts [Reference Hilbe and Hilbe30].
Person-time incidence rates for E. coli O157 infection over the 8-year study period per 100 000 Alberta population in each division level were standardized using the age structure of the 2006 Alberta population. Average incidence rates for the study period were calculated and reported for the CDRS and DAD data separately. Separate monthly standardized incidence rates for the CDRS and DAD data were also calculated for each CD. Empirical Bayes smoothing was applied to the standardized rates and analysed separately [Reference Clayton and Kaldor31]. Thus, CDRS and DAD datasets were each separated into three sets of rates. Incidence rates for the 8-year study period at the CD level and the CCS level, and monthly incidence rates at the CD level. Standardized incidence rates smoothed via empirical Bayes were also considered. HUS was also of particular interest as this outcome of E. coli O157 can result in death. Standardized rates of HUS by CD from the DAD dataset were also analysed. Standardized incidence rates were examined for autocorrelation by observing Moran's I statistic [Reference Haining and Haining32].
Multicollinearity in model covariates can result in unwarranted increases to standard errors, thus potentially deeming significant variables insignificant. To deal with this issue correlations between all covariate pairs was first tested. If the Pearson product-moment correlation coefficient was >0·8, covariates were segregated into one of five different groups. The degree of multicollinearity for each group was determined via a variance inflation factor (VIF), which indicates how much the variance was increased due to collinearity in the model's covariates. The VIF was calculated for each initial group of covariates and covariates moved between groups such that a minimum VIF was achieved in all groups. The first models fitted to the data consisted of covariates from one of these five groups. A stepwise model selection procedure based on Akaike's Information Criterion (AIC) was then run on each model and significant covariates regrouped into additional models and again tested via the stepwise AIC procedure. The final model suggested by the stepwise AIC procedure was then examined for significant regression coefficients. Coefficients were considered significant if the P value was < 0·05. Covariates having regression coefficients deemed non-significant were then removed from the model. The remaining covariates were also removed from the model if their regression coefficients were deemed non-significant when one of the other significant variables was removed from the model. Final models were compared taking into consideration model fit, parsimony, interpretability, AIC and Bayesian Information Criterion (BIC). Models were fitted in R version 2.15.1 and spatial procedures were performed in OpenGeoDa. Model fit was assessed using the global Moran's I test for autocorrelation of regression residuals, Breusch–Pagan test for homoscedastic residuals, robust Jarque–Bera test for normally distributed errors, residual diagnostic plots, and in the case of the count models, overdispersion tests and goodness-of-fit tests on the residual deviance were also applied [Reference Anselin29].
RESULTS
Geographical and spatial outcomes
The geographical distribution of standardized incidence rates of E. coli O157 cases from the CDRS data is presented in Figure 1. The highest CDRS rates of E. coli O157 occurred in the southern regions of Alberta, with the largest incidence rates coming from CD 2, 3 and 4 (12·81, 14·52 and 11·75 cases/100 000 person-years, respectively). The CCS having the highest CDRS rate of E. coli O157 (22·8 cases/100 000 person-years) was Pincher Creek; this location is indicated by an asterisk in Figure 1. The lowest CD for CDRS standardized rates of E. coli O157 was from divisions 14 and 17 each having <1·3 cases/100 000 person-years.

Fig. 1. Standardized incidence rates for Communicable Disease Reporting System cases of E. coli O157 in Alberta, 2004–2011 (cases/100 000 person-years). Numbers on the map indicate census division numbers. * Pincher Creek census consolidated subdivision.
The highest DAD rates occurred in CD 4, 7 and 17. The DAD standardized rates were lowest in CD 6 and 11.
Spatial autocorrelation was found to be present in standardized incidence rates for CDRS data at both the CD and CCS levels as Moran's I statistic was significant regardless of underlying null distribution (P < 0·00 566). The DAD data presented no spatial autocorrelation at the CD level but the CCS level showed highly significant spatial autocorrelation regardless of underlying null distribution (P < 1·657 × 10−7) in both the standardized and empirical Bayes smoothed rates.
Model fit
Count models provided superior fit to that of the OLS, spatial lag and error models. Overdispersion was often found in the Poisson and Poisson zero-inflated regression models but the equivalent negative binomial models were able to account for the greater variability. The spatial error and lag models fitted well but the spatial terms were often not statistically significant and thus, according to parsimony, an OLS procedure would be considered a better fit.
Collinearity
Collinearity in the livestock variables was very high with the bull, calf and beef cow variables being highly correlated. Bull:person, beef:person, calf:person and calf:child ratios each had Pearson product-moment correlation coefficients >0·993. Similarly, bull:area, beef:area and calf:area ratios had correlation coefficients >0·949. Weather variables of similar type also had a high degree of collinearity. Temperature variables had correlation coefficients between 0·8645 and 0·95 44 while precipitation and rain variables had coefficients between 0·8612 and 0·9498.
Significant risk factors
Covariates found significant in models for the CDRS data included calf:person ratio, cattle density quartiles, CD type, latitude, longitude, season, mean temperature, cumulative rainfall, horse density and deer density. The final best-fitting models and their significant risk factors for the CDRS data are given in Table 3. Calf:person ratio was highlighted as a significant variable in all final models presented. Bull:person, beef:person and calf:child ratios were also found to be highly significant in final models; however, since these variables are so highly correlated with each other only calf:person ratio was included in the final model, as it led to slightly decreased AIC values. This decrease in AIC value was generally less than a single unit, thus models fitted using the covariates bull:person, beef:person or calf:child ratios could be of equal standing. All models estimated that a larger calf:person ratio would increase the expected number of E. coli O157 infections when all other risk factors are held constant. A lower latitude in Alberta was also estimated to have an increased number of E. coli O157 infections by all final models when all other risk factors are held constant.
Table 3. Best-fitting models for the CDRS data. Models presented were selected based on proper model fit and having the lowest AIC values
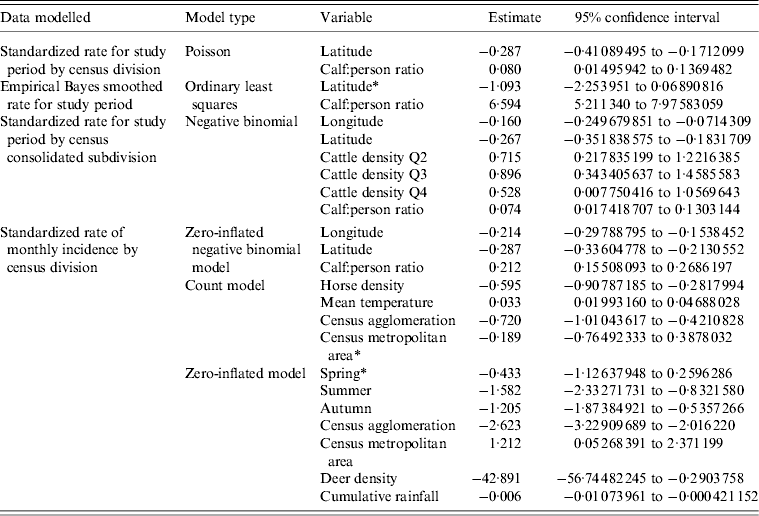
CDRS, Communicable Disease Reporting System; AIC, Akaike's Information Criterion.
* Variables are not statistically significant at the 0·05 significance level.
CD type, CCS type, longitude, latitude, mean temperature, sheep density, calf:person ratio and beef:person ratio were determined to be significant covariates in final models for the DAD data. Final best-fitting models and their significant risk factors for the DAD data are given in Table 4. The spatial distribution of calf:person ratio can be seen in Figure 2. All models fitted using the DAD data estimated that a larger calf:person ratio would increase the expected number of E. coli O157 infections when all other risk factors are held constant. Areas deemed to be census agglomeration or census metropolitan had a lower number of expected E. coli O157 infections compared to their rural counterparts when all other risk factors were held constant.

Fig. 2. Average calf:person ratio (total number of calves per census division/total human population of census division) in Alberta, 2004–2011. Numbers on the map indicate census division number.
Table 4. Best fitting models for the DAD data. Models presented were selected based on proper model fit and having the lowest AIC values
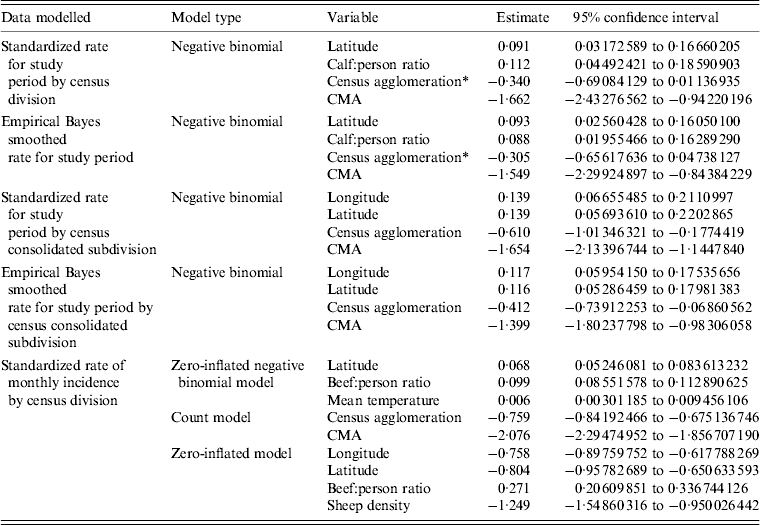
DAD, Discharge abstract database; AIC, Akaike's Information Criterion; CMA, census metropolitan area.
* Variables are not statistically significant at the 0·05 significance level.
HUS analysis
A significant spatial autocorrelation was found in the HUS standardized incidence rates as Moran's I statistic had a P value of 0·0425. Count models were not used on this data as rounding could misconstrue results due to the low incidence rate during this study period. An ordinary least squares model was identified as the best-fitting model, as spatial terms were not significant in the respective spatial models. Latitude and calf:person ratio were identified as significant risk factors in the OLS model.
DISCUSSION
The results of this analysis provide evidence of a statistically significant association between human E. coli O157:H7 infection/HUS and measures of cattle density in the province of Alberta, Canada. CDRS cases have less misclassification of E. coli O157 disease, as they are all culture confirmed and do not include other (non-O157) E. coli cases as do the DAD cases. Spatial misclassification of geographical areas as having ‘high’ or ‘low’ human disease rates may also result from differential access to health services or other elements of the surveillance system. The spatial distribution of E. coli O157 as depicted by the CDRS data is consistent with Alberta Health and Wellness' communicable disease report [1] presenting a subset of the CDRS data used in the current study.
High incidence rates of E. coli O157 tend to exist in the southern regions; however, DAD data identified CD 17 as having a high incidence rate. This high incidence rate is the result of a large number of gastritis cases. As noted in the Methods section, the DAD data were included in this study to act as a sensitive E. coli O157 dataset. It is unknown whether cases observed in the DAD data were actually E. coli O157 infections; thus, for example some cases of gastritis may in fact be the result of causes other than E. coli O157. CD 17 is unique in that it has the lowest population density along with the highest proportion of people who identify themselves as Aboriginal, i.e. 39% [33]. Pearl et al. [Reference Pearl15] investigated socioeconomic risk factors for rates of E. coli O157 in Alberta using CDRS data and showed a sparing effect for areas with a high Aboriginal population (>10%). The sparing effect in areas highly populated by Aboriginals was hypothesized by Pearl et al. [Reference Pearl15] to have been the result of differences in diet, environmental exposures and requirements for disease reporting. Furthermore, Goodman et al. [Reference Goodman34] presented research showing increased susceptibility of Aboriginal populations to Helicobacter pylori, a major cause of gastritis. The large Aboriginal population in CD 17 could result in a low E. coli O157 incidence rate in the CDRS data because of the sparing effect described by Pearl et al. [Reference Pearl15]; while a high E. coli O157 incidence rate in the DAD data was observed because of the increased number of gastritis cases.
Measures of cattle density, such as calf:child, calf:person, beef:person or bull:person ratios have been shown to be associated with human E. coli O157 in Alberta. This evidence, as well as confirmation from multiple studies in multiple countries, suggests that the cattle–human E. coli O157/HUS association is valid. The calf:person ratio was highly correlated with values for beef:person, bull:person and calf:child ratios, each being highly significant in the model. Calf:child ratio was found to have an association with incidence of paediatric HUS in France [Reference Haus-Cheymol27]. The ratio of beef cattle to human population was also found to be a significant predictor of shiga toxin-producing E. coli infection in Ontario [Reference Valcour10].
Spatial autocorrelation of standardized incidence rates was observed for the CDRS data; this was also observed in an Ontario study of verocytotoxigenic E. coli conducted by Michel et al. [Reference Michel9]. Pearl et al. [Reference Pearl35] also studied spatial association of E. coli O157 in Alberta using spatial scan statistics and found spatial clusters in southern Alberta that varied slightly by year. In the DAD data, spatial autocorrelation was only detected at the CCS level; this may suggest that when more E. coli O157 cases are observed and underreporting is not present, the spatial association is based on a smaller division level than the CD. The repeated significance of latitude and longitude in the majority of the best-fitting models also point to a spatial component of E. coli O157 infection. Despite taking various potential risk factors into account, latitude may be considered as a significant variable due to confounding with a risk factor not studied. The gradual increase in population from the northern to southern regions of Alberta may illuminate socioeconomic or demographic factors acting as said confounder. In addition, potential changes in land use and climate could also account for latitude being a potential confounding variable.
No livestock variables, other than a select few cattle variables, were found significant in models selected via AIC criteria of the average E. coli O157 incidence rates. Horse density and deer density were identified as significant model parameters of the zero-inflated negative binomial model (ZINB) for monthly incidence of the CDRS data by CD; along with sheep density in the DAD analogous version of this model. Increasing horse density is estimated to decrease the number of human E. coli O157 infections. Increases in deer and/or sheep density are estimated to decrease the odds of a zero monthly incidence rate being an excessive zero. Thus, there is a higher probability that in areas of high deer and/or sheep density a zero monthly incidence rate is a true zero and not due to non-reporting. CD with high horse density generally had low calf:person ratios. Similarly, areas of high deer and/or sheep density were associated with low calf:person ratios. Competition for area resources is a potential explanation of this inverse phenomenon. High horse density according to the model is indicative of lower expected incidence values. This protective effect may be due to the absence of cattle in areas of high horse density. Beef:person ratio in the DAD ZINB model and calf:person ratio in the CDRS ZINB models for monthly incidence rate by CD are highly correlated with each other and were identified as increasing the expected incidence count. The lack of evidence supporting other livestock associations with E. coli O157 supports a true cattle–human illness association; the association seen makes biological sense and is consistent with the literature.
Weather variables were not found to be significant in models selected via AIC criteria of incidence rates based on the full study period for both the DAD and CDRS data. Mean annual temperature and median annual precipitation of districts were also found not to be significant in a study of E. coli O157:H7 attributed HUS cases in children of France [Reference Haus-Cheymol27]. The spatial and temporal spread used for the full study period models may not be adequate in differentiating weather variable significance. Monthly incidence rates of E. coli O157 for both the CDRS and DAD data had final models including mean temperature as a significant predictor, which increased the incidence count in the ZINB model. Cumulative rainfall was also acknowledged as being a significant risk factor in increasing the odds that an observed incidence rate of zero was an excessive zero. This indicates that increased rainfall in a given month and region is less likely to have a zero incidence rate observed be a true zero incidence rate. This result corresponds with earlier studies that suggested rainfall, extreme rainfall in particular, and high temperature, in the form of a degree-days variable, increased the odds of a waterborne disease outbreak [Reference Thomas13, Reference Curriero16]. Season was also included in the zero-inflated portion of the ZINB model for the CDRS monthly incidence model, with summer and autumn seasons associated with 79% and 70% lower odds of having a true zero incidence compared to winter, this corresponds correctly with E. coli O157's peak season [Reference Michel9]. Weather information collected on a smaller spatial and temporal scale could provide superior data and covariate analysis; however, the weather variables included in the ZINB models do show evidence that temperature and rainfall are important risk factors in E. coli O157 spread; however, further research is required.
CD and CCS type were significant risk factors according to the DAD data models. A difference in expected incidence rates between the rural and CMA was consistently found with the CMA divisions having significantly lower incidence rates when holding all other covariates fixed, this was also seen in other studies in Ontario [Reference Michel9, Reference Valcour10]. Monthly incidence ZINB models of both the DAD and CDRS data identified CD type in the count model portion and the CDRS data also identified CD type in the zero-inflated portion of the model. Pearl et al. [Reference Pearl15] also identified that statistical area classification was an important risk factor when modelling E. coli O157 in Alberta at the CSD level and noted that rural, or CSD not influenced by metropolitan areas, had greater rates of disease.
The degree of aggregation of results allows for the detection of small increases in risk in the population but care must be taken when interpreting the results as ecological fallacy can occur when making causal inferences at an individual level [Reference Morgenstern36]. Risk factors identified in this paper may be helpful in considering community-level disease spread of E. coli O157. A causal relationship between these risk factors and disease incidence cannot be established solely on the basis of this investigation due to limitations in the present study design.
Results of CDRS data analysis in this study thus provide evidence of a significant association between measures of cattle density in Alberta and human E. coli O157 disease. This is true under multiple different modelling scenarios and is supported by the association between DAD measures of human illness and cattle density measures, again using multiple different modelling approaches. It is also supported by the association between cattle density measures and HUS, in spite of low observation numbers. The source of increased human E. coli O157 illness in areas of high cattle density is most likely due to direct contact with cattle, cattle manure and drinking water (e.g. for the farm wells) contaminated with cattle manure. Effective measures aimed at reducing the risk associated with direct and indirect exposure of rural residents in the province to cattle manure are warranted to address this issue.
ACKNOWLEDGEMENTS
This work was supported by funding provided by Bioniche Life Sciences Inc. The authors acknowledge the support of the Ontario Ministry of Agriculture Foods and Rural Affairs (OMAFRA) through the OMAFRA – University of Guelph Highly Qualified Personnel Graduate Scholarship to Nadia Bifolchi. Thanks are also due to Alessia Guthrie for all her hard work in helping to organize and keep this project on track.
DECLARATION OF INTEREST
None.











