Cryptosporidium species are the most common protozoan parasites in Scotland affecting both immunocompetent and immunocompromised individuals. The symptoms of cryptosporidiosis include diarrhoea, abdominal pain, nausea, vomiting, malaise and weight loss which occur due to the ingestion of infective oocysts. Sporozoites released from oocysts attach to, and invade the intestinal mucosa resulting in significant morbidity and even mortality in the immunocompromised. Cryptosporidiosis is endemic worldwide and transmission can be anthroponotic or zoonotic with spread occurring by faecal–oral, food and waterborne routes [Reference Bouzid1].
Cryptosporidium hominis is the principal cause of anthroponotic cryptosporidiosis in many regions including the USA, Europe and Australia and is thought to account for about half of the annual reported cases of human cryptosporidiosis in England and Wales [Reference Xiao2–Reference Wielinga5]. It has a narrow host tropism and principally infects humans with laboratory-confirmed cases of animal infections being relatively uncommon.
Improvements in public health reporting systems and molecular technologies allow greater understanding of the biology and epidemiology of Cryptosporidium species, particularly those associated with outbreaks or significant disease at a national level. Since April 2012, data pertaining to the molecular diversity of C. hominis in Scotland has been generated as part of the remit of the Scottish Parasite Diagnostic and Reference Laboratory (SPDRL). Diagnostic bacteriology laboratories from every health board in Scotland routinely analyse human faeces for the presence of Cryptosporidium oocysts using microscopy. It is not possible to differentiate Cryptosporidium species by this method, therefore molecular technologies are employed. Only those microscopy-positive faeces which are deemed to be from potential outbreaks are forwarded to SPDRL for molecular analysis. The definitions used by Health Protection Scotland to describe an outbreak are (a) an incident in which two or more linked cases experience the same illness or (b) where the observed number of cases unaccountably exceeds the expected number.
Faeces which were positive for Cryptosporidium oocysts using microscopy were sent to SPDRL for molecular analysis only if they were suspected of being part of an outbreak. Faeces were subjected to water/ether concentration and the sporozoite DNA extracted using the QIAamp DNA Stool Mini kit incorporating a 10-min incubation step of 95 °C using the manufacturer's ASL buffer (Qiagen, Germany). Speciation was performed using real-time PCR assays [Reference Hadfield6] while a nested PCR approach was used to subtype samples by targeting the glycoprotein-60 gene (GP60) [Reference Glaberman7, Reference Sulaiman8]. PCR-positive samples were subjected to bi-directional sequencing (Applied Biosystems 3500XL) and the EMBI and CBI Blastn website tools used to search for sequence similarities. Subtypes were confirmed by manually reading through the sequences to search for trinucleotide repeats and other repeat sequences [Reference Xiao2].
A total of 1139 microscopy-confirmed individual cryptosporidiosis cases were observed during 2012 (n = 710) and 2013 (n = 429) (Table 1). Duplicate and/or follow-up samples were omitted from the analysis. Speciation was performed in 445 samples (39% of the total), with 187, i.e. 42·0% of those speciated shown to be C. hominis (n = 129, 2012; n = 58, 2013). Of the remaining isolates that were speciated, the most common to be identified was C. parvum (n = 256, 57·5%). These findings are comparable with Scottish data from previous years where the percentage of C. hominis cases ranged from 33·3% to 53·1% during 2006–2010 while the percentage of C. parvum cases ranged from 43·0% to 59% over the same time period [Reference Chalmers9].
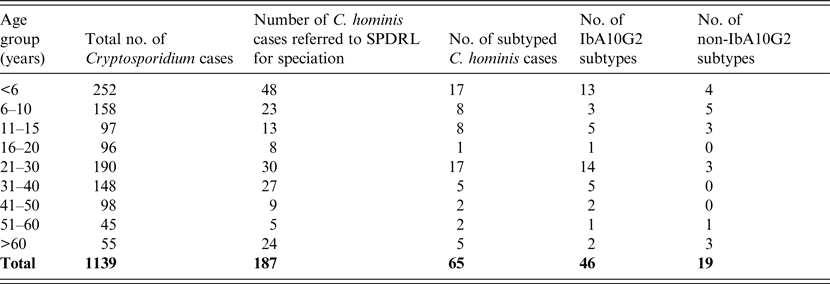
Table 1. Age distribution and subtypes of C. hominis infected individuals residing in Scotland during 2012 and 2013
SPDRL, Scottish Parasite Diagnostic and Reference Laboratory.
During 2012 and 2013, one case of C. felis and one case of C. meleagridis were also identified.
These data highlight a 40% reduction in cases during 2013 compared to 2012. There have been no changes to standard laboratory procedures for identifying and reporting Cryptosporidium species that could account for this reduction. Extreme prolonged low temperatures occurred in Scotland during late 2012/early 2013 and this may have reduced the viability of oocysts in the environment resulting in fewer cases during 2013. However, it should be noted that the numbers of cases reported to Health Protection Scotland in 2012 (n = 710) were unusually high compared to 2007–2011 (5-year average n = 567). Contributing factors which may account for the high levels during 2012 include two widespread UK/European outbreaks of cryptosporidiosis, one involving cases of C. parvum, and the other C. hominis. The C. parvum outbreak occurred during May 2012, with salad leaves implicated as the most likely source which resulted in an increased awareness of this disease [Reference McKerr10]. In addition, during late summer of that same year, an increase in the number of cases was observed not only in the UK, but also in The Netherlands and Germany involving C. hominis with no single common source identified [Reference Fournet11].
Almost half of the total number of Cryptosporidium infections identified by microscopy were from individuals aged ⩽15 years (n = 507, 45%; Table 1). Similarly, of the 187 isolates that were shown to be C. hominis by molecular analysis, 45% (n = 84) were within this same age group (Table 1). Of the 65 cases that were subjected to subtyping, the majority (n = 33, 50·8%) were also from patients aged ⩽15 years followed by the 21–30 years age group (n = 17, 26·2%) (Table 1). These data are likely to reflect either person-to-person contact or the presence of a common source during social interactions between parents and children.
A total of 103 females were infected with C. hominis and 81 males (sex not stated, n = 3). This may reflect the preference of the different sexes for certain social activities or perhaps the greater likelihood of females seeking medical attention and submitting samples.
There was marked seasonality with a clear surge in the number of isolates occurring during October/November in both years corresponding to the autumn/winter season where about half of all C. hominis isolates occurred during this period (Fig. 1) (2012: n = 66, 51%; 2013: n = 30, 52%). This is a similar finding to Scottish data from previous years [Reference Chalmers9].
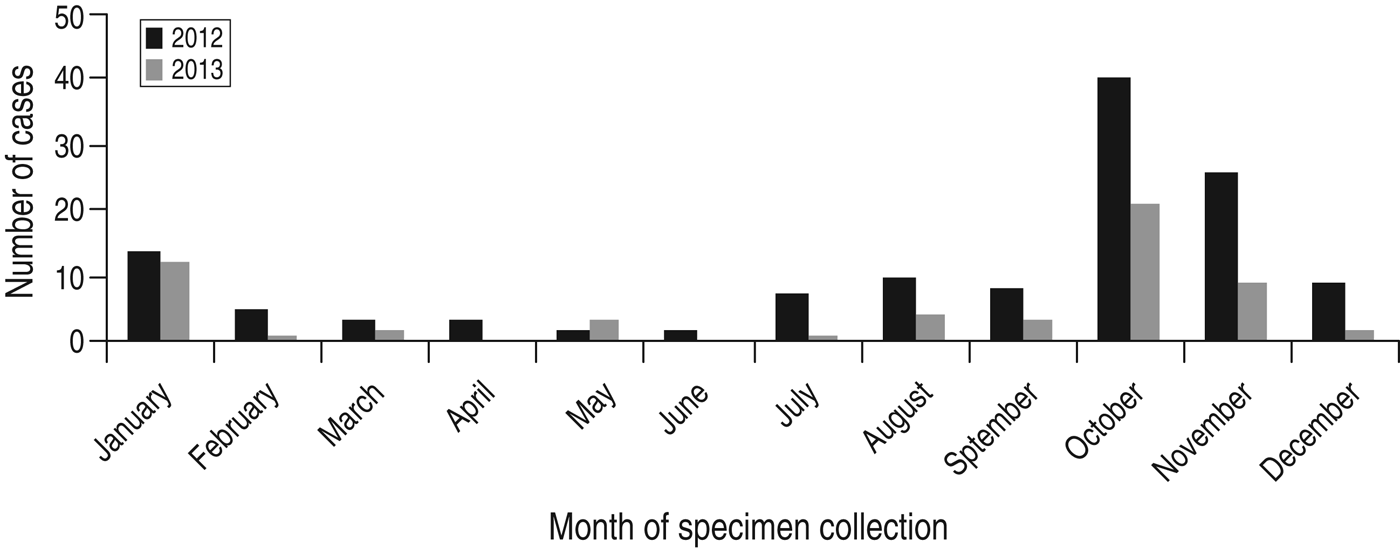
Fig. 1. Seasonal distribution of C. hominis isolates collected from human cases residing in Scotland.
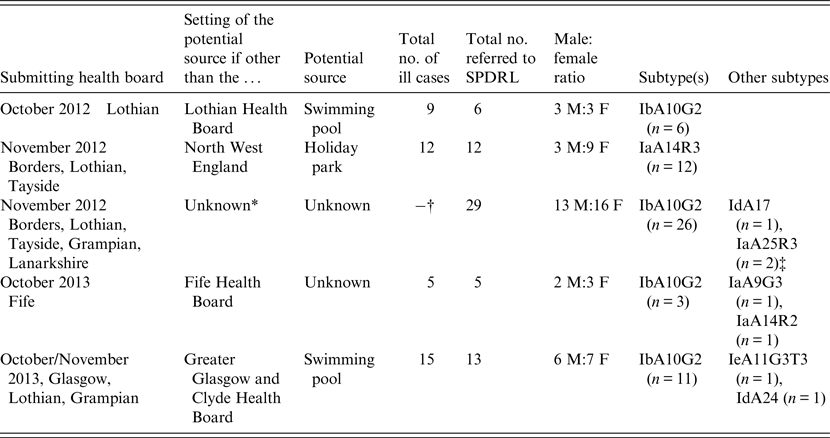
The 65 C. hominis isolates that were subjected to subtyping having been deemed to be part of potential outbreaks were all from cases residing in Scotland; however, in only two of the five outbreak investigations were the outbreak settings known to be from within Scotland (Table 2). In one of the other outbreaks, all cases were resident within Fife at the time of the investigation but the potential source of infection was not identified. In another outbreak, investigations involved residents from Scotland; however, the outbreak setting was located in the North West of England, specifically at a holiday park (Table 2). No supporting evidence was available to suggest a potential link to any one particular source within the holiday park. The largest Scottish outbreak investigation of 2012 occurred during autumn (Table 2) in line with similar increases in The Netherlands, Germany and England [Reference Fournet11]. The remainder of C. hominis cases were not subjected to subtyping in response to further information from Health Protection Scotland and local public health bodies which indicated they were unlikely to be linked to other cases.
Table 2. Summary of outbreak investigations involving C. hominis IbA10G2 and non-IbA10G2 subtypes isolated from Scottish residents
SPDRL, Scottish Parasite Diagnostic and Reference Laboratory.
* In addition to increases of Cryptosporidium infections in Scotland, similar increases were also noted in the rest of the UK, The Netherlands and Germany [Reference Fournet11].
† A total of 66 cases of Cryptosporidium infections were reported in Scotland during October/November 2012. No single source was identified.
‡ Two related infants. The mother of both cases was suspected of having cryptosporidiosis but no sample was submitted for confirmation.
Contaminated swimming pools were implicated in two of the Scottish outbreaks (Table 2) which is not surprising as robust oocysts can be resistant to treatment with chlorine. Swimming pool-associated Cryptosporidium infections have been reported worldwide, and the resistance of oocysts to chlorine combined with the direct interactions of individuals, particularly children, increases the risk of cryptosporidiosis in these settings [Reference Stafford12, Reference Polgreen13].
Although there can be a predominance of C. hominis in urbanized areas which are more densely populated [Reference Pollock14], in this study, C. hominis was isolated from humans located in a broad range of geographical areas including both urban and rural Scottish regions. Of the 65 samples that were subtyped, the majority of isolates were submitted from the two largest Scottish health-board regions, Lothian (n = 21, 32%) and Glasgow (n = 13, 20%) (Fig. 2). Other health board regions throughout Scotland that submitted samples for subtyping included the Borders, Ayrshire, Tayside, Lanarkshire, Grampian, and Fife (Fig. 2).
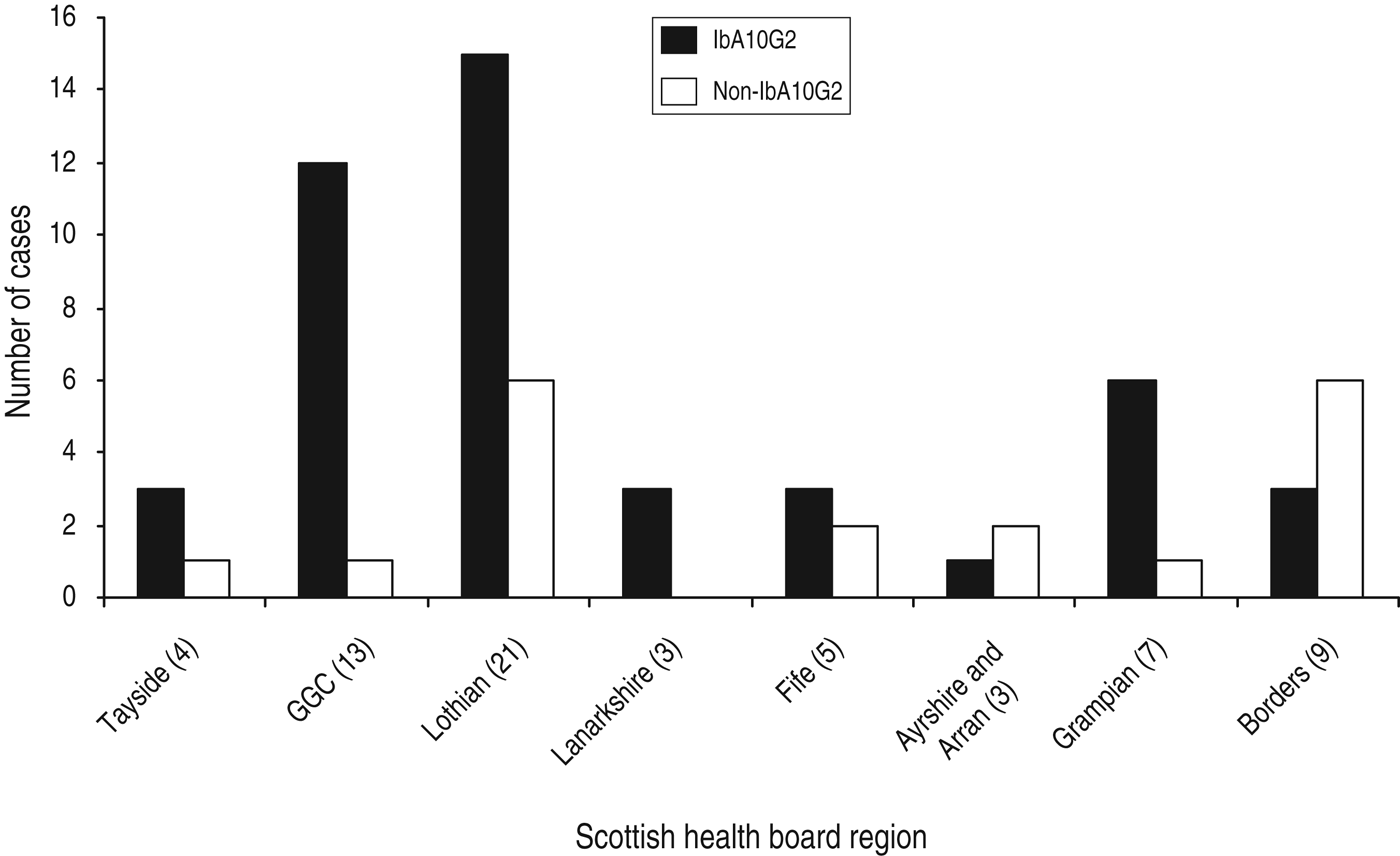
Fig. 2. Distribution of IbA10G2 and non-IbA10G2 C.hominis isolates within the Scottish health board that was home residence for each cryptosporidiosis case during specimen collection.
The most prevalent subtype identified was IbA10G2, which comprised 71% (n = 46) of all those investigated (Tables 1 and 2). The Ib family is associated with outbreaks worldwide and this particular subtype is one of the commonest in Western countries [Reference Xiao2]. This subtype accounts for about half of all C. hominis outbreaks in the USA [Reference Xiao2] and, in addition, a study covering Wales and North West England demonstrated that 80% of C. hominis isolates from sporadic cases were of this subtype [Reference Chalmers15]. Its identification in a large number of Scottish isolates may reflect its increased virulence compared to other subtypes which is likely to encourage those infected to seek medical advice. Of the non-IbA10G2 subtypes, IaA14R3 was the most common in Scottish outbreak investigations, having been identified in 12 cases from three Scottish regions where all cases were likely to have been infected at the same setting in the North West of England (Table 2).
Other, less common subtypes isolated from Scottish cases were identified as part of outbreak investigations (Table 2). Although there were common exposure(s) to initially suggest they may have been part of a specific outbreak, due to having a subtype that differed from the predominant subtype, another (unidentified) source may have been implicated. However, the possibility of a mixed infection where different sub-populations exist within a single host must also be considered. Four isolates from the Ia family were noted, IaA14R2 (n = 1), IaA9G3 (n = 1), IaA25R3 (n = 2), and two from the Id family, IdA24 (n = 1) and IdA17 (n = 1). Both the Ia and the Id family have been isolated from human cases outside the UK including India, Ireland and Kuwait [Reference Sulaiman8, Reference Gatei16–Reference Zintl18]. Information highlighting imported infections is often excluded from laboratory request forms and only in nine of the 445 samples submitted to SPDRL was there reference to recent foreign travel. Despite there being no record of recent travel histories for the Scottish cases infected with these less common Ia and Id family subtypes, there remains the possibility that these isolates were imported.
Cama et al. have reported in children, that first infections caused by the Ia family, but not further infections, were associated with a greater number of symptoms including vomiting and nausea [Reference Cama19]. Gathering further information on future Scottish cases to include detailed clinical symptoms and duration/frequency of episodes would permit comparisons with previously published data.
One Scottish case had an unusual C. hominis isolate from the Ie family, namely IeA11G3T3. It is not known if the Scottish case had any recent foreign travel history to regions where this particular subtype has been isolated from environmental and human sources to explain this unusual finding. These include China, the Gulf of Guinea, Mexico, India, Australia and Kuwait [Reference Sulaiman8, Reference Gatei16, Reference Feng20–Reference Waldron23]. An interesting report from England demonstrated an association of non-IbA10G2 subtypes with foreign travel [Reference Chalmers15] but as enhanced surveillance of cryptosporidiosis in Scotland is not performed, it is not possible to state with certainty if international travel was a factor in every non-IbA10G2 case. However, it is known that in 13 of the 19 non-IbA10G2 cases, no foreign travel was implicated. For the remaining six cases, no travel details were provided.
The subtyping of isolates has provided a valuable insight into the diversity of C. hominis within Scotland and their geographical distribution. The introduction of enhanced surveillance of cryptosporidiosis in Scotland is essential to provide crucial in-depth data to assist with future outbreak investigations. In addition, further studies are required to examine novel biomarkers to increase our understanding of the molecular complexity of this parasite and the virulence of specific subtypes in humans.
ACKNOWLEDGEMENTS
We thank Lisa Connelly at the Scottish Parasite Diagnostic and Reference Laboratory for her technical support. This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
DECLARATION OF INTEREST
None.









