INTRODUCTION
Members of the genus Bartonella are aerobic Gram-negative bacteria which are mainly vector-borne and recognized as emerging pathogens [Reference Boulouis1]. Many new species or subspecies have been described in recent years and the genus currently consists of at least 30 species, 16 of which are associated with human diseases [Reference Chomel2]. The clinical spectrum of symptoms in humans caused by Bartonella species is considered to be widening [Reference Boulouis1]. The most common infection is cat scratch disease, caused by B. henselae, with domestic cats (Felis catus) being the natural reservoir [Reference Regnery, Martin and Olson3–Reference Abbott5]. Domestic cats are also the natural reservoir for B. clarridgeiae and B. koehlerae [Reference Kordick6–Reference Yamamoto8].
Free-ranging wild felids, just like domestic cats, can be infected with Bartonella species [Reference Chomel9–Reference Chomel11]. In North America, free-ranging mountain lions and bobcats are infected with Bartonella species similar to but different from B. henselae and B. koehlerae, proposed as B. koehlerae subsp. boulouisii and B. koehlerae subsp. bothieri [Reference Chomel11, Reference Chomel12], and also infected with B. henselae [Reference Chomel12, Reference Girard13]. In Africa, B. henselae was isolated from free-ranging lions (Panthera leo) from Kruger Park, South Africa [Reference Molia10]. It was also isolated from semi-captive lions from three ranches in the Free State Province, South Africa [Reference Pretorius14] and Kelly et al. [Reference Kelly15] had reported the isolation of B. henselae from a pet cheetah in Zimbabwe. A ftsZ gene sequence close (99%) to B. koehlerae has also been detected by polymerase chain reaction (PCR) in a captive margay cat (Leopardus wiedii) born in the wild and kept at Sao Paulo Zoo [Reference Filoni16].
As we have recently described new subspecies of B. koehlerae in wild felids from North America [Reference Chomel12], the objective of this study aimed to fully describe the Bartonella species previously isolated from free-ranging cheetahs from Namibia and lions from Kruger Park, South Africa [Reference Molia10] in order to determine if they belonged to a new Bartonella species or subspecies, as well as Bartonella isolated from captive cheetahs at the San Diego Zoo Safari Park, CA, USA.
MATERIALS AND METHODS
Bacterial strains
B. henselae type I (Houston 1) ATCC 49882 and B. clarridgeiae ATCC 51734 were obtained from the American Type Culture Collection (Rockville, MD). Strain U4 originally isolated from a naturally infected cat at the University of California Davis was used for the B. henselae type II (Marseille type). B. koehlerae strain C29 (ATCC 700693) was isolated from a naturally infected barn cat [Reference Droz7]. Candidatus B. weissii (now B. bovis), isolated from domestic cats and the B. henselae Zimbabwean cheetah strain were kindly provided by Russell L. Regnery (Centers for Disease Control and Prevention, USA).
Clinical samples
Animals. Whole blood samples (1·5–2·0 ml per tube) were collected in EDTA plastic tubes (Becton Dickinson and Company, USA) from 58 free-ranging lions at Kruger Park, South Africa and 17 cheetahs held in a rehabilitation centre after being rescued from various regions of Namibia, as previously reported [Reference Molia10]. At the San Diego Zoo Safari Park, blood samples were collected from 11 cheetahs in 1999 (Table 1), of which four had blood collected more than once. These animals were kept in a large open enclosure during the day. Collection of blood samples by veterinarians was performed under animal use and care protocols of the San Diego Zoo Safari Park and at the Cheetah Conservation Fund facility. Animals were tranquillized using 11 mg/kg ketamine hydrochloride via the intramuscular route. All blood samples were stored at −20 °C or lower until they were processed for culture.
Table 1. Bartonella bacteraemia level, serological titres, sex, age and date of collection of blood samples from 11 semi-captive cheetahs (Acinonyx jubatus), San Diego Zoo Safari Park, California

B.h., Bartonella henselae; c.f.u./ml, colony-forming units/millilitre; IFA, immunofluorescence assay.
Blood culture
In 2003, the whole-blood samples were defrosted and then centrifuged at 5000 g for 30 min at room temperature. Blood pellets were resuspended in 125 µl M199 inoculation medium [Reference Koehler17] and plated onto heart infusion agar (Difco Laboratories, USA) containing 5% fresh rabbit blood. The plates were then incubated at 35°C in 5% CO2 for 4 weeks, and cultures were examined at least twice a week for bacterial growth. The number of colonies observed was recorded as the number of colony-forming units per milliliter (c.f.u./ml) of blood. Colonies were subcultured, harvested, and frozen at −70 °C.
Microscopic analysis
Gram staining was performed on all isolates. Sterile swabs moistened with filter-sterilized phosphate-buffered saline (PBS) were used to remove colonies from subcultured agar plates to glass slides. Following heat fixation, Gram staining was used for light microscopic visualization of organisms. The motility of cells suspended in heart infusion broth was determined with a 100 × oil immersion objective.
Indirect immunofluorescence antibody (IFA) test
Antibody titres against B. henselae were determined using an IFA test [Reference Molia10]. For antigen preparation, B. henselae (strain U4) was cultivated on Vero cells for 3–5 days. Infected Vero cells were then suspended in L15/MEM tissue culture medium (Invitrogen, USA). Infected Vero cells were applied to each well of multi-well super-cured heavy Teflon-coated slides (Erie Scientific Co., USA) and incubated for 24 h to allow the cells to adhere to the slides. Slides were then rinsed in PBS (pH 7·4), air-dried, acetone-fixed, and stored at −20 °C after a final air-drying. Wild cat sera (supernatant from the whole-blood samples) were diluted 1:64 in PBS with 5% skim milk, and added to the test wells of the slides. After washing in PBS, fluorescein-labelled goat anti-cat IgG (Cappel, Organon Teknika Corp., USA) was used as the conjugate. Positive and negative controls were included on each test slide. Intensity of fluorescence of the bacteria was graded subjectively on a scale of 0–4. Fluorescence intensity ⩾2 at a dilution of 1:64 was considered to be a qualitatively positive result, as described previously [Reference Chomel9]. For a quantitative result, positive sera were serially diluted (twofold dilutions) to obtain an endpoint titre. Reading of the slides was performed independently by two readers for all serum samples.
DNA extraction and PCR
Subcultured colonies were scraped off and suspended in 100 µl sterile water. The bacterial suspension was heated for 15 min at 100 °C and centrifuged at 15 000 g for 10 min at 4 °C. The supernatant, diluted 1:10, was then used as a template for amplification of the 16S rRNA, gltA, ribC, groEL and ftsZ genes as well as the 16S-23S intergenic spacer region (ITS). About 380 base pairs (bp) of the gltA gene [Reference Norman18], 1500 bp of the 16S rRNA gene [Reference Gurfield19], 1500 bp of the groEL gene [Reference Marston, Sumner and Regnery20], 700 bp of 16S-23S ITS [Reference Roux and Raoult21], 764 bp of the ftsZ gene [Reference Zeaiter, Liang and Raoult22], 586 bp of the ribC gene [Reference Johnson23] and fragments of various size of 16S-23S ITS [Reference Jensen24] were amplified using previously described primers and methods and verified by gel electrophoresis.
Restriction fragment length polymorphism (RFLP)
The amplified products were enzymatically digested overnight using various restriction endonucleases. The amplified product of the gltA gene was digested with TaqI (Promega, USA), HhaI, MseI, and AciI (New England Biolabs, USA), the amplified product of the 16S rRNA gene was digested with DdeI (New England Biolabs), the amplified product of the ribC gene was digested with TaqI, and the amplified product of the 16S-23S ITS gene was digested with TaqI and HaeIII. The temperature of digestion was 65 °C for TaqI and 37 °C for all other enzymes. The digested fragments were separated by electrophoresis in a 3% Nusieve GTG agarose gel (Biowhittaker Molecular Applications, USA). Fragment sizes were estimated by comparison with a 100-bp ladder (Invitrogen).
Sequencing
PCR products used for DNA sequencing were purified with QIAquick PCR purification kit (Qiagen Sciences, USA) and sequencing was done in both directions using a fluorescence-based automated sequencing system (Davis Sequencing, USA). For the phylogenetic analysis, sequence data of the gltA, ftsZ, rpoB [Reference Renesto25] genes and the ITS region were imported into Vector NTI Suite 9.0 software (Invitrogen) in order to obtain a consensus sequence. Align X in Vector NTI was utilized for aligning sequence variants with each other and other known species of Bartonella for each of the genes or ITS. A neighbour-joining tree was constructed using MEGA v. 5.0 (http://www.megasoftware.net) by concatenating the four sequences [Reference Tamura26]. Bootstrap replicates were performed to estimate node reliability of the phylogenetic tree, with values obtained from 1000 randomly selected samples of the aligned sequence data. The evolutionary distances were computed using the Kimura two-parameter method [Reference Kimura27].
RESULTS
Primary isolation
In primary isolation, very small colonies were observed growing on 5% rabbit blood heart infusion agar plates after at least 14 days following plating of the blood pellets from three (5·2%) of the 58 lions, one (5·9%) of the 17 Namibian cheetahs, as previously reported [Reference Molia10] and eight (73%) of the 11 semi-captive cheetahs from California (Table 1). All bacterial colonies were homogeneous, either rough or smooth, round, grey-white in colour, 0·3–1·0 mm in diameter and embedded in the medium. The numbers of c.f.u./ml for the lion's isolates were respectively 35 and 93 c.f.u./ml in two adult females and 2000 c.f.u./ml in a juvenile male, as previously reported [Reference Molia10]. For the adult male cheetah that was culture positive, the number of colonies was 49 c.f.u./ml. For the Californian semi-captive cheetahs, bacteraemia ranged between 5 and 1322 c.f.u./ml (Table 1).
Microscopic examination.
Microscopic examination after Gram staining revealed short, slender Gram-negative rods. No motility or flagella were detected.
Serology
Serological results for the African free-ranging wild felids have been presented previously [Reference Molia10]. For the semi-captive cheetahs from California, five (45·5%) animals were seropositive with titres ranging from 1:64 to 1:128. Two cheetahs had low titres (⩽64) and five culture-positive cheetahs were seronegative.
PCR/RFLP analysis
Two of the three lion isolates had profiles similar to B. henselae for the gltA gene when using TaqI, HhaI and AciI enzymes and the ITS gene using TaqI. The PCR/RFLP profiles after digestion of the gltA gene by TaqI and HhaI endonucleases showed that the Namibian cheetah strain (1178) and the lion strain 98–215 were similar to B. koehlerae subsp. koehlerae's profile with only lion strain 98–215 having a profile similar to B. koehlerae subsp. koehlerae after digestion with AciI. A unique profile for cheetah strain 1178 and lion strain 98–215 was obtained for the gltA gene using HhaI endonuclease (Fig. 1), and for the ribC gene using TaqI endonuclease (data not shown), different from all other wild cat strains, but similar to B. koehlerae and B. bovis (ex weissii). Using the DdeI enzyme for the 16S rRNA gene, all isolates were determined to have the same profile as B. henselae type II, with the exception of the lion isolate 98-215 which was similar to the profiles of B. henselae type I, B. koehlerae subsp. koehlerae and B. bovis (formerly B. weissii).

Fig. 1. PCR/RFLP of the gltA gene using HhaI endonuclease, showing a unique profile for cheetah strain 1178 and lion strain 98–215 compared to all other wild felid isolates (no digestion), similar to B. koehlerae and B. bovis (ex weissii).
Sequencing
As previously reported, B. henselae was isolated from the blood of two of the bacteraemic lions (isolates 150·41 and 7069). Both isolates were identified as B. henselae type II (Marseille) based on partial sequencing of the 16s RNA gene. The third isolate from a juvenile (aged <24 months) male lion (98-215) was determined to be 100% identical to B. koehlerae subsp. koehlerae by partial sequencing of three different genes (gltA, ftsZ, rpoB) and the 16S-23S ITS, as shown for the concatenated tree (Fig. 2). The three bacteraemic lions were from different locations in Kruger National Park, South Africa. The Bartonella strain isolated from the blood of the bacteraemic cheetah from Gobabis District, Namibia was a previously unidentified strain temporarily designated as ‘Cheetah 1178’. The California captive cheetahs all had an identical PCR/RFLP profile and partial sequencing revealed that they were identical to B. koehlerae subsp. bothieri (strain L1796), isolated from a California bobcat [Reference Chomel12].
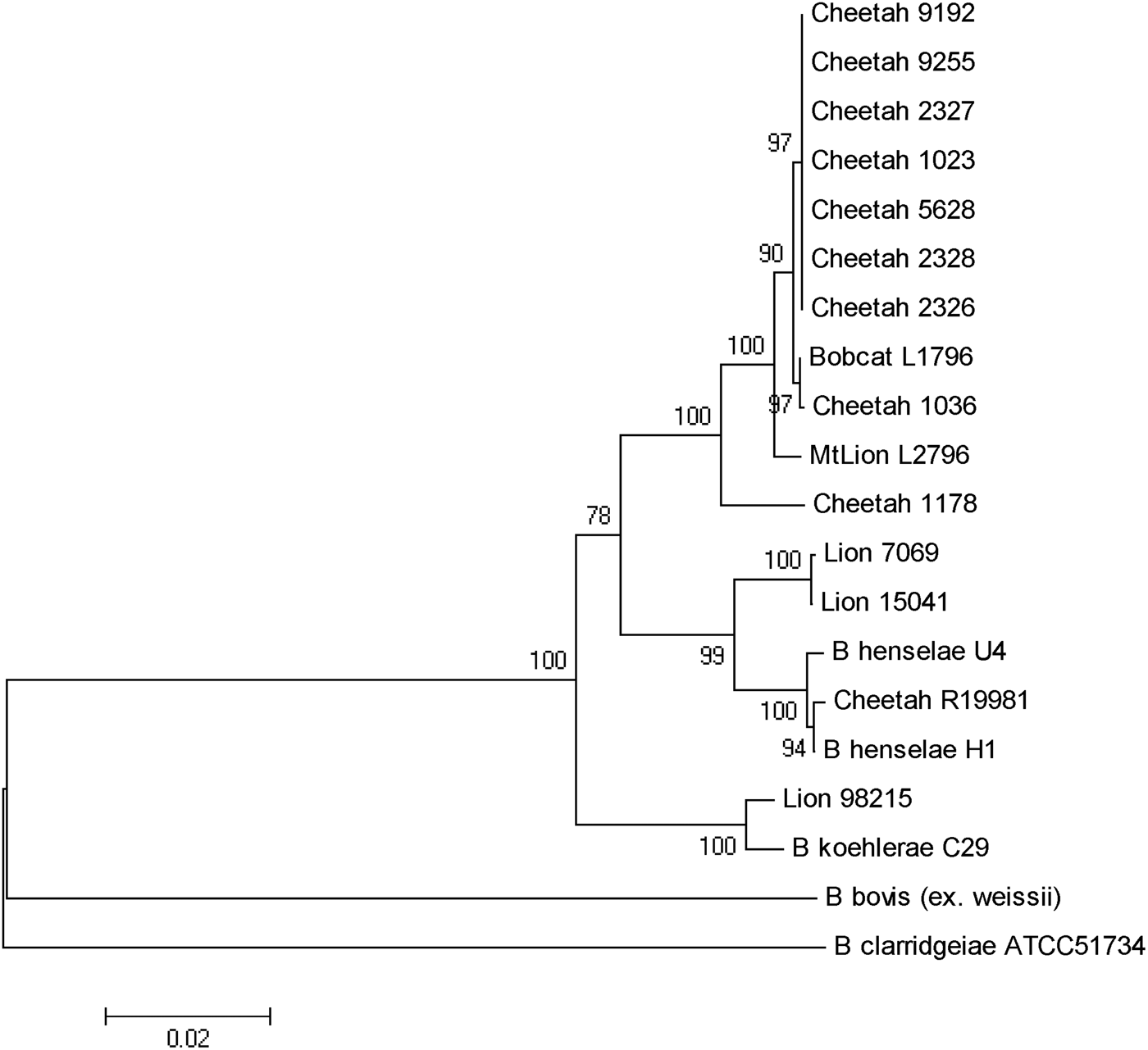
Fig. 2. Phylogenetic tree of Bartonella species based on the combined gltA, rpoB, ftsZ, and intergenic transcribed spacer sequence alignment. The tree shown is a neighbour-joining tree based on the Kimura two-parameter model of nucleotide substitution. Bootstrap values are based on 1000 replicates. The analysis provided tree topology only; the lengths of the vertical and horizontal lines are not significant.
DISCUSSION
B. henselae was isolated from two adult lions, as reported previously [Reference Molia10]. Both isolates were identified as type II (Marseille) based on partial sequencing of the 16s RNA gene. An isolate of B. henselae type II was also reported from a lion living in one of three game farms in the Free State, South Africa [Reference Pretorius14]. The third isolate from a young male lion was determined to be 100% identical to B. koehlerae subsp. koehlerae by PCR/RFLP and by partial sequencing of two different genes (gltA ftsZ) and the 16S-23S ITS. This is the first report of isolation of B. koehlerae subsp. koehlerae from a free-ranging wild felid and expands the spectrum of felids that can be naturally infected with this Bartonella species. Until that isolate, B. koehlerae subsp. koehlerae had only been isolated from domestic cats in the United States, France and Israel [Reference Droz7, Reference Rolain28–Reference Gutiérrez, Nachum-Biala and Harrus30] and B. koehlerae subsp. koehlerae DNA had been detected from cat and rodent fleas [Reference Rolain28, Reference Marié31], a captive margay cat [Reference Filoni16] as well as from three of 61 dogs that were culture- or PCR-positive for Bartonella species after enrichment [Reference Pérez32]. It was also identified in two cases (a human and a dog) of endocarditis [Reference Avidor29, Reference Ohad33]. This study confirms that besides domestic cats, free-ranging wild felids are infected and could be reservoirs of B. henselae, the agent of cat scratch disease, and B. koehlerae subsp. koehlerae, a known human pathogen.
Kelly et al. [Reference Kelly15] reported the isolation of B. henselae genotype II from a pet cheetah from Zimbabwe. The present study is the first to report the isolation of a Bartonella species from a wild-caught cheetah. The PCR/RFLP profiles, using various endonucleases, led to a specific profile (no digestion) for cheetah strain 1178 and lion strain 98–215 for the gltA gene using HhaI endonuclease and for the ribC gene using TaqI endonuclease. This was different from all other wild cat strains but similar to B. koehlerae and B. bovis. Furthermore, partial sequencing of gltA, groEL, ftsZ genes and ITS put this Namibian cheetah strain (1178) in an intermediate position between B. henselae and B. koehlerae, as shown for strains isolated from free-ranging mountain lions and bobcats from California [Reference Chomel12], but clearly distinct from the cheetah isolates from the San Diego Zoo.
The high infection rate of the semi-captive cheetahs from southern California was unexpected, but could be explained by the frequent contacts between these animals and the sharing of ectoparasites such as fleas. Interestingly, all the cheetah isolates were different from B. henselae or B. koehlerae subsp. koehlerae isolates, and from the Namibian cheetah isolate, clustering with the B. koehlerae subsp. bothieri strains isolated from free-ranging bobcats (Lynx rufus) in California. They also were distinct from the B. koehlerae subsp. boulouisii strain isolates from mountain lions (Felis concolor) from California. Therefore, it is very likely that the semi-captive cheetahs became infected by an indigenous strain for which bobcats are the natural reservoir, possibly through infected fleas carried by bacteraemic bobcats.
The strains isolated from mountain lions, bobcats and cheetahs were morphologically similar to B. henselae and B. koehlerae, but they were not noticeable on blood agar for 2 weeks after plating. By contrast, Bartonella colonies are usually visible within a few days for domestic cat isolates. This characteristic was also observed in specific pathogen-free domestic kittens experimentally infected with one of the mountain lion isolates [Reference Yamamoto8].
ACKNOWLEDGEMENTS
The authors thank Russell L. Regnery for providing the cheetah strain from Zimbabwe. This project has been funded in part by the George and Phyllis Miller Feline Research Fund, Center for Companion Animal Health, U.C. Davis, California, the Master of Preventive Veterinary Medicine Research Project Fund (University of California, Davis) and Mérial Inc., Athens, GA. Sophie Molia was a recipient of a Lavoisier grant (French Ministry of Foreign Affairs) and Barron fellowship (University of California, Davis). Jane E. Koehler received funding support from a California HIV Research Program Award, and NIH grants U54AI065359 and R01AI103299 from the NIAID.
DECLARATION OF INTEREST
None.








