INTRODUCTION
Mumps virus is a non-segmented, single-stranded, negative-sense RNA virus of the Paramyxoviridae family. The infection is initiated by contact of the viral surface glycoprotein haemagglutinin-neuraminidase (HN) with the receptor on the host cell surface. Thus, HN is a major target of humoral immunity in mumps virus infection. Classification of mumps virus isolates has been updated recently [1]. Mumps virus is classified in 12 genotypes designated A–N (genotypes E and M were merged with genotypes C and K, respectively) based on the sequence diversity of the small hydrophobic (SH) protein.
Mumps virus causes disease which is characterized by acute parotitis, fever, headache and lethargy. In some cases it can cause aseptic meningitis, encephalitis, pancreatitis and orchitis as severe complications which may lead to permanent sequelae. Many countries have implemented the vaccine against mumps in their routine vaccination programmes, mainly as MMR (measles-mumps-rubella) vaccine. The two-dose vaccination schedule is believed to provide long-term immunity. However, recent resurgence of mumps in doubly vaccinated cohorts has been observed, mostly identifying viruses of genotype G as the cause of epidemics [Reference Watson-Creed2–Reference Walker10]. Assumed reasons for ongoing mumps outbreaks included primary- or secondary-vaccine failure (reviewed in [Reference Peltola11]). Furthermore, a limited vaccine efficacy might be due to the incomplete cross-neutralization between vaccine strain and a circulating wild-type (WT) virus which is somewhat antigenically different (reviewed in [Reference Peltola11]). Cases of mumps re-infection have been described [Reference Gut12–Reference Yoshida14] and were usually ascribed to antigenic diversity between virus causing primary infection and the re-infecting virus. The currently used vaccine strains originate from the mid-20th century and are phylogenetically distant from currently circulating viruses. The purpose of this work was to question the capacity of antibodies raised upon immunization with the three most used vaccine strains to neutralize WT strains isolated during 1998–2011, some of which belong to genotype G, which seems to be the predominant genotype currently.
MATERIALS AND METHODS
Mumps virus strains
Mumps virus strains used in this study were: L-Zagreb (Institute of Immunology), Urabe AM9 (1st International Reference Reagent for Mumps Vaccine, NIBSC), JL5 (a kind gift from B. K. Rima), 9218/Zg98 [Reference Šantak15], Du/CRO05 [Reference Šantak15], Zg/CRO06 [Reference Ivancic-Jelecki, Santak and Forcic16] and MuVi/Split.CRO/05.11 (isolated in Croatia in 2011). Further details regarding the virus strains are given in Table 1. All viruses were propagated in Vero cells (African green monkey kidney cells; ATCC) for up to three passages. Vero cells were maintained in minimal essential medium with Hank's salts (MEM-H) supplemented with 10% fetal calf serum (FCS) and 50 μ g/ml neomycin. Viruses were grown at 35 °C in medium with 2% FCS until cytophatic effect was observed.
Table 1. Mumps virus strains used for immunizations of guinea pigs and as antigens in serological assays

* Vaccine strain or wild-type strain (type of case).
Virus titre was determined by both haemagglutination and plaque assay. Haemagglutination inhibition (HI) was performed according to Mahy & Kangro [Reference Mahy, Kangro, Mahy and Kangro20], with exception that 0·5% guinea pig erythrocyte suspension was used. Plaque-forming units (p.f.u.) were determined according to Forcic et al. [Reference Forcic21].
Immunization of guinea pigs
All animal work was in accordance with Croatian Law on Animal Welfare (2006) which strictly complies with EC Directive (86/609/EC). Guinea pigs of CRL:(HA)BR strain, bred at the Institute of Immunology, were used for immunization experiments. Antigen for immunization was prepared by ultracentrifugation of mumps virus suspensions at 104 000 g in a SW28 rotor (Beckman Coulter, USA) for 2 h and the pellet was resuspended in PBS (pH 7·0). Five animals (two females, three males) were immunized with each vaccine strain. Animals were given three subcutaneous immunizations (days 0, 21, 35) each containing 80 haemagglutination units. Forty-two days following prime immunization, guinea pigs' blood was collected by heart puncture and sera were prepared for further serological analyses by centrifugation at 1200 g at 4 °C for 30 min and decomplementation by heating at 56 °C for 30 min.
Serological methods
HI was performed according to Cross [Reference Cross22], with exception that 0·5% guinea pig erythrocyte suspension was used.
Plaque-reduction neutralization test was performed as in Cohen et al. [Reference Cohen23], with some modifications. Test sera were prepared in six fourfold dilutions starting from at least one dilution completely neutralizing the added virus, and ending with at least one dilution having no neutralizing capacity. Viruses to be neutralized are listed in Table 1. The virus concentration was adjusted to 250 ± 50 p.f.u./ml with MEM-H with 2% FCS. Testing was performed in 24-well plates in which Vero cells were seeded at a concentration 1 × 105 cells/well 48 h prior the addition of mixture serum/virus. Each dilution of the serum was mixed with an equal volume of virus and incubated at 35 °C for 1 h. Then the medium was removed from the plate and 0·2 ml mixture was transferred into four wells in the plate. Each assay run included controls: virus as positive control, medium for diluting the sera, and viruses as negative control. The plates were incubated at 35 °C for 1 h, the mixture serum/virus was removed and the cells were overlayed with 0·6 ml semisolid medium consisting of 1 (v/v) 2 × MEM-H with 10% FCS without Phenol Red and 1 (v/v) 1·5% Noble agar (Sigma). Plates were incubated at 35 °C in a humidified atmosphere of 5% CO2. After 6 days the monolayers were stained with 1 ml of 0·05% Neutral Red (Sigma) and plaques were counted. Fifty percent endpoint titres were calculated using the Kärber formula: log10 ND50 = − max − Δ(∑p − 0·5), where max is the log10 of the highest dilution of serum in the test, Δ is log10 of the dilution factor (for fourfold dilution is −0·6) and ∑p is total number of plaques divided by the number of plaques in positive control sample.
Phylogenetic analysis
For phylogenetic analysis a complete SH gene sequence (6218–6534, 316 nt) was used. A deduced amino acid (aa) sequence (582 aa) of complete HN gene was used for phylogenetic analysis of antigenic relatedness between vaccine and WT strains.
Sequences included in phylogenetic analyses were (accession numbers given in parentheses): Urabe AM9 (X99040), L-Zagreb (AJ272363), JL5 (AF338106), MuVi/Boston.USA/0.45 (U980052), MuVi/Nottingham.GBR/19.04 (JQ034452), MuVi/Shandong.CHN/4.05 (EU780221), MuVi/Gloucester.GBR/32.96 (AFF280799), MuVi/Bedford.GBR/0.89 (JQ945273), MuVi/Akita.JPN/42.93 (JQ945274), MuVi/Sapporo.JPN/12.00 (AB105475), MuVi/RW154.USA/0.70s (JQ945276), MuVi/Fukuoka.JPN/41.00 (AB105483), MuVi/Taylor.GBR/0.50s (AF142774), MuVi/Tokyo.JPN/0.93 (AB003415), MuVi/London.GBR/3.02 (AY380077), 9218/Zg98 (EU370206), Zg/CRO06 (EU259202, EU259203), ES06-ABx1 (AM420295), ES06-ADx1 (AM420297), MuVs-GBR0300796 (EU597478), MuV-IA (JN012242), MUM/NewJersey.US/2006 (DQ661744), MuVi/Split.CRO/05.11 (JN635498), Du/CRO05 (EU370207), MuVs-DEU10-18695 (FR687016), MuVs/Mass/19.09 (GU056309), ES06-AFx1 (AM420299), Wrek1/UK97 (AF142775), UK00-46x7 (AY380073), Kent/UK97 (AF142768), UK99-177x2 (AY380067), UK99-162x3 (AY380066), UK02-302 (AY380076), UK00-322x2 (AY380072), UK01-4x23 (AY380074), UK00-47x4 (AY380074), UK99-208x22 (AY380069), MuVs-GBR00-25827 (EU597477), UK01-22 (AY380075), SA856/Ja01 (AB085231), SA841/Ja00 (AB056141), SA702/Ja99 (AB056145), MuVs-USA06-NH923 (FJ959107), MuVi/Nottingham.GBR/19.04 (JQ034464), MuVi/Tokyo.JPN/6.01 (JQ946043), MuVi/Shandong.CHN/4.05 (JQ034463), MuVi/Sheffield.GBR/1.05 (JQ946046), MuVi/Sapporo.JPN/12.00 (JQ946044), MuVi/London.GBR/3.02 (JQ946038), MuVi/Calgary.CAN/30.07 (JN687469), MuVi-BRA07-2484-M (JF268685).
All alignments were performed by Clone Manager Suite software (Scientific & Educational Software, USA). Unrooted neighbour-joining (NJ) trees were generated by ClustalX software (University College Dublin) and visualized with NJ plot [Reference Perrière and Gouy24].
RESULTS
Phylogenetic analysis
Survey of the scientific literature (see http://www.ncbi.nlm.nih.gov/sites/entrez) for the number of publications on the topic of the mumps outbreaks or associated vaccine failure studies in the period 2000–2011 revealed 154 publication in English (Fig. 1). The number of publications was constant (2–3 publications per year) until 2004 when the number begins to rise. In 2007 it reaches a peak with 29 publications, and then the number decreases to around 20 publications per year. A detailed search of the literature indicated that genotype G became the predominant mumps virus genotype. Therefore a number of mumps virus isolates belonging to genotype G which are accessible through the GenBank database were included in phylogenetic analysis. To examine phylogenetic relationships between them and to follow their evolution, representative strains in terms of location and time of isolation were included in this study. A phylogenetic analysis based on the SH gene of vaccine and WT strains used in this study was performed and the phylogenetic tree is shown in Figure 2. The study focused on sequences which represented a complete SH gene while shorter sequences were excluded. Their limited size required downsizing of all sequence lengths which would influence the reliability of results obtained by phylogenetic analyses. Figure 2 shows the phylogenetic tree indicating two clusters in genotype G. One cluster (labelled as Japan, USA 1999–2006) contained a single branch of Japanese isolates. The exception to the Japanese strains was strain MuVs-USA06-NH923 which was isolated during an outbreak in Virginia [Reference Rota4] and was not linked to the large mumps outbreak in Midwestern USA that occurred earlier in 2006. Thus this strain might have been imported from Japan and had its own direction of evolution. The second cluster represents strains which clearly originated in the UK in the mid-1990s. Between that time and 2002 isolates reported to GenBank were mainly from the UK (labelled UK 1996–2002) except for above-mentioned Japanese strains. Later years show the outgrowth of a branch of isolates from the USA and different European countries (Spain, UK, Croatia, Germany) (labelled Europe, USA 2003–2011).

Fig. 1. The number of publications on mumps outbreaks or associated vaccine failure studies during 2000–2011.
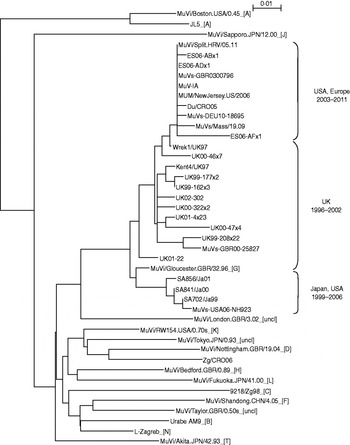
Fig. 2. Phylogenetic tree based on the entire SH gene presenting currently assigned reference strains (genotypes are indicated by the capital letters in square brackets; uncl, unclassified strains) [1], three vaccine strains (L-Zagreb, Urabe AM9, JL5), representative strains of genotype G (including strains Du/CRO05 and MuVi/Split.CRO/05.11) and two wild-type viruses of non-genotype G (9218/Zg98 and Zg/CRO05).
Strains Du/CRO05 and MuVi/Split.CRO/05.11 were isolated in Croatia in 2005 and 2011, respectively. Both viruses caused a limited outbreak, the first in 2005 [Reference Šantak15] and the second in 2011. Both of these strains belong to genotype G, branch USA, Europe 2003–2010 (Fig. 1). Although isolated 5 years apart, phylogenetically they are very close to each other with only 1 nt difference in the SH gene while they differ at 66 nt positions throughout the genome. The phylogenetic analysis indicates that isolate MuVi/Split.CRO/05.11 is identical to isolates MUM/NewJersey.US/2006, MuV-IA, MuVs-GBR0300796 and ES06-ADx1. However, comparison of complete genomes shows that Du/CRO05 and MuV-IA are more similar to each other (38 nt difference) although they seem to be more distant in the phylogenetic tree than MuVi/Split.CRO/05.11 and MuV-IA which show a 54 nt difference at the level of the whole genome. This indicates that when performing detailed analyses, a fragment of the SH gene might not be long enough, while for rough screening it may be sufficient.
Three vaccine strains L-Zagreb, Urabe AM9 and JL5 are separately located in the tree: L-Zagreb has been recently designated as genotype N [1]. Urabe AM9 is a reference strain for genotype B and JL5 is positioned in genotype A, whose branch is remote from the rest of the tree. Strains 9218/Zg98 and Zg/CRO06 were isolated also in Croatia as single cases imported from abroad, the first in 1998 [Reference Šantak15] and the second in 2006 [Reference Ivancic-Jelecki, Santak and Forcic16]. Phylogenetically they are distant from genotype G strains (Fig. 2): the 9218/Zg98 belongs to genotype C and Zg/CRO06 belongs to genotype D.
Analyses of the mumps virus neutralizing antibodies in guinea pig sera
The main purpose of this study was to examine the capacity of antibodies raised against the three most used mumps vaccine viruses, L-Zagreb, Urabe AM9 and JL5, to neutralize two WT strains of genotype G and also two strains of non-genotype G. The four WT strains were isolated in a 13-year period.
Genotyping based on the SH (presented above), although informative, cannot lead to a conclusion on the cross-reactive potential between different mumps viruses. To study this issue five guinea pigs were immunized with each of the vaccine strains and collected sera were initially analysed by HI against the virus which was used for immunization. All 15 sera were seropositive with titres between 12 800 and 102 400. Since the correlation of serological test results and clinical protection is particularly poor in mumps vaccination [Reference Peltola11, Reference Plotkin25, Reference Allwinn26], two types of tests were used in this study: the HI test and the plaque reduction neutralization test (PRNT). Each serum was tested with both methods against seven mumps viruses: three vaccine strains used for immunizations and four WT strains described above.
The HI and PRNT titres were comparable as shown in Figures 3 and 4, respectively. All sera were able to neutralize either strain although at different levels. Sera raised against all three vaccine strains were able to neutralize the other two vaccine strain viruses at the same level, except JL5, which was slightly less efficient in the HI test. When neutralizing WT viruses, the neutralization efficiency of sera decreases several fold by temporal order, meaning that the viruses which are isolated earlier in time are more susceptible to neutralization with the vaccine-induced sera. This may indicate that gradual evolution of mumps viruses, rather than belonging to a certain genotype, is a major cause of the decrease in neutralizing efficacy of guinea pig sera immunized with vaccine strains.

Fig. 3. Serum levels of mumps virus antibodies in guinea pig sera immunized with either (a) L-Zagreb, (b) Urabe AM9, or (c) JL5 measured by haemagglutination inhibition (HI) test. •, Represents the titre value of a single serum; ◊, represents the average value of all sera.

Fig. 4. Serum levels of mumps virus antibodies in guinea pig sera immunized with either (a) L-Zagreb, (b) Urabe AM9, or (c) JL5 measured by plaque-reduction neutralization test (PRNT). •, Represents the titre value of a single serum; ◊, represents the average value of all sera.
Analysis of the HN epitopes
The HN protein is the major target of neutralizing antibodies. Therefore, the HN protein of virus strains used in this study were compared and analysed.
Phylogenetic analysis based on the previously established HN-specific epitopes [Reference Kovamees27–Reference Cusi29] was initially performed (data not shown). However, since there are no differences in these regions of the HN protein between L-Zagreb, Urabe AM9, Du/CRO05 and MuVi/Split.CRO/05.11 this phylogenetic tree might lead to false conclusions. Therefore the phylogenetic tree based on the complete HN protein was drawn and is depicted in Figure 5. The phylogenetic tree (Fig. 5) shows that JL5 appears at a separate branch from the others. L-Zagreb and Urabe AM9 and are more similar to each other than to the other strains. Strains Du/CRO05, Zg/CRO06 and MuVi/Split.CRO/05.11 are further separated indicating their similarity (especially Du/CRO05 and MuVi/Split.CRO/05.11 which differ at three amino-acid positions). Strain 9218/Zg98 is positioned at a separate branch. The result of the similarity between the HN proteins of the viruses used is in agreement with the neutralization test results described above. Several sequences of the HN protein of contemporary WT mumps virus strains from GenBank were included in the analysis (Fig. 5) to further confirm the hypothesis.

Fig. 5. Phylogenetic tree based on the complete HN protein sequence presenting three vaccine strains (L-Zagreb, Urabe AM9, JL5) and several wild-type strains during 1998–2011.
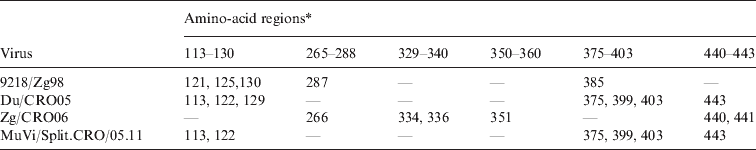
The finding that the known epitopes did not show any difference between L-Zagreb, Urabe AM9, Du/CRO05 and MuVi/Split.CRO/05.11 suggested that the antibodies were directed against other areas of the protein representing epitopes relevant for virus neutralization. In order to identify potential epitope sequences, an alignment of the putative HN protein sequence was performed. The comparison of the cytosolic and transmembrane region of the HN protein (1–55 aa) reveals that these regions are not highly conserved regions since the incidence of amino-acid differences to the vaccine viruses ranges between 3·6% (9218/Zg98) and 12·7% (Du/CRO05 and MuVi/Split.CRO/05.11). However, neutralization relevant epitopes are located in the extracellular part of the HN protein which is far more conserved with the incidence of amino-acid differences compared to the vaccine viruses being between 1·3% (MuVi/Split.CRO/05.11) and 2·0% (Zg/CRO06). The protein sequence alignment shows that most of the differences between vaccine strains and WT strains are grouped in six regions which are summarized in Table 2. Three of them were previously described (265–288, 329–340, 350–360) (Table 2, Fig. 6a) as relevant epitopes [Reference Kovamees27–Reference Cusi29] and the other three are newly identified potential epitopes in this study (113–130, 375–403 440–443) (Table 2, Fig. 6b). Only Zg/CRO06 strain has dominant escape neutralization epitopes in regions 265–288, 329–340 and 350–360, and two additional differences in region 440–443, which may have an additional effect on the neutralization efficacy of antibodies induced by vaccine strains. The regions 113–130, 375–403 and 440–443 may play a dominant role in the other three WT strains. There are additional differences compared to vaccine strains at positions 76 and 362 for 9218/Zg98, at position 153 for Du/CRO05 and at positions 59, 74, 410 and 511 for Zg/CRO06. Interestingly, the MuVi/Split.CRO/05.11 strain does not have any additional difference to the vaccine strains while being the strain least affected by the neutralizing antibodies, which supports the relevance of the herein identified epitopes. The sequence alignment analysis also included the WT strains included in the phylogenetic analysis shown in Figure 5. The sequence alignment (Fig. 6) further confirms that these regions are potentially relevant and should be further studied.

Fig. 6. Pairwise sequence alignment of the previously identified (a) epitopes and (b) regions suggested in this study to be involved in the viral escape from neutralization antibodies for the HN protein of the three mumps virus vaccine strains (L-Zagreb, Urabe AM9, JL5) and several wild-type mumps virus strains. Genotypes are indicated by capital letters in parentheses. Amino acids differing from the L-Zagreb strain are indicated by single-letter codes.
Table 2. The positions of amino acids in the three epitope regions (265–288, 329–340, 350–360) and three potential epitope regions (113–130, 375–403, 440–443) of the HN protein where four wild-type mumps viruses differ from the vaccine strains L-Zagreb, Urabe AM9 and JL5
* Only amino-acid positions of the extracellular part of the HN protein are shown.
DISCUSSION
It has been shown more than once that the two-dose vaccine regimen against mumps is beneficial (reviewed in [Reference Peltola11]). The receipt of one dose leaves the vaccinee in danger because chances to receive natural boosters are continuously lessening. However, the number of reports of mumps outbreaks in doubly vaccinated cohorts is increasing (Fig. 1). Most of the outbreaks are caused by mumps viruses belonging to genotype G [Reference Watson-Creed2–Reference Walker10]. It appears that genotype G is now endemic in Ireland and the UK [Reference Carr6]. The nature of these outbreaks raises questions regarding the efficacy of the mumps virus vaccine in each of these cohorts. Primary- or secondary vaccine failure has been suggested as the cause of the outbreaks. Mumps virus is unlikely to exist as multiple serotypes since the range of HN amino-acid identity in all published sequences is 93·84–100% [Reference Rubin30]. However, the ineffectiveness of the vaccine against heterologous genotypes has also been considered [Reference Nöjd13, Reference Örvell28, Reference Yates, Afzal and Minor31].
Mumps virus of G genotype was initially isolated in the UK in 1996. Since then until 2002 it spread mostly in the UK (except for a branch with Japanese stains, Fig. 2) although the low level of notifications from other sites might lead to a false conclusion. Nevertheless, it appears that the evolution of genotype G began in the UK and then (season 2002/2003) spread throughout North America and the rest of Europe (Fig. 2). The evolution might have been driven by the interplay of two promoters: the vaccine-induced protection was strong enough to create a bottleneck but not strong enough to completely suppress the virus that was aided by the fact that MMR coverage in the UK began to fall during the 1996/1997 season [Reference Kay32]. All this created an opening for WT viruses to escape from vaccine-induced immunity.
Although guinea pigs are not susceptible to infection with mumps virus, they show an abundant immunological response to the live virus (reviewed in [Reference Cantell33] and [Reference Flynn, Mahon and Pandalai34]) and are widely used as a small animal model for testing of immunity provoked by live mumps vaccines. This study analysed the capacity of guinea pig sera immunized with the three most used vaccine strains, L-Zagreb, Urabe AM9 and JL5, to neutralize WT viruses and addresses the question whether genotype G is the genotype that was able to escape to vaccine-induced immunity or is the gradual evolution of any mumps virus, the potential factor that can be responsible for vaccine failure. Vaccine strains are of different genotypes (see Fig. 2) but all three vaccine strains originated from viruses isolated in the 1950s (L-Zagreb) or 1960s (Urabe AM9, JL5) (Table 1) which is more than 40 years of mumps virus evolution.
Neutralization capacity of human sera (either from vaccinees or after exposure to WT mumps virus) to neutralize mumps strains Enders (genotype A) and Lo1 (genotype B) was compared by Rubin et al. [Reference Rubin30]. Nearly all sera tested were able to neutralize both virus strains. However, significant differences in neutralization titres were observed. The latter confirms differences in antigenicity in mumps virus strains. The findings in this work support the findings of Rubin et al. [Reference Rubin30] and, furthermore, show that more significant differences in neutralization titres were observed the more that the WT strain is temporally distant from the vaccine strains. This also supports the results of the phylogenetic analysis of the HN gene presented in Figure 5. The highest difference measured by HI test was 16-fold (JL5 vs. MuVi/Split.CRO/05.11, Fig. 3) and measured by PRNT was 18·6-fold (Urabe AM9 vs. MuVi/Split.CRO/05.11, Fig. 4). Human sera either from vaccinees or from individuals who were exposed to natural infection with mumps virus typically have low anti-mumps antibody titres (the highest HI titre reached was 1600; our unpublished data). With the level of reduction of neutralization efficacy observed herein for WT strains, the anti-mumps antibody titres will be decreased even further.
HN protein is a major target of humoral immunity. So far there are three regions (265–288, 329–340, 350–360) known to be antigenic [Reference Kovamees27–Reference Cusi29]. Comparison of sequences of HN proteins of the seven mumps virus strains identified three new regions (113–130, 375–403, 440–443, Fig. 6b) potentially relevant for the escape from the neutralization of strains 9218/Zg98, Du/CRO05 and MuVi/Split.CRO/05.11 by the guinea pig sera immunized by the vaccine strains. Epitopes identified earlier together with the newly identified region 440–443 seem to be responsible for the escape from neutralization of Zg/CRO06 by the vaccine strains. Interestingly, virus MuVi/Split.CRO/05.11 is one amino acid less different from the vaccine strains than Du/CRO05 but is more resistant to neutralization by vaccine-induced antibodies, indicating that the neutralization process involved is a more complex mechanism, probably involving other viral proteins such as fusion proteins.
In conclusion, a vaccine failure in two-dose-vaccinated cohorts seems to be the result of incomplete neutralization of WT viruses by the antibodies raised by vaccine immunization which decrease over the years [Reference Dayan3, Reference Date35, Reference Snijders36]. To overcome this problem a more effective vaccine should be developed or a new vaccine policy involving a third dose should be considered by the national authorities.
Acknowledgements
The authors are grateful to B. K. Rima for providing JL5 mumps virus strain. R. Mažuran and M. Markušić are acknowledged for constructive discussion. This work was supported by the Ministry of Science, Education and Sports of the Republic of Croatia (grant number 021-0212432-3123 to M.Š.).
DECLARATION of interest
The authors (except S.Lj-S) are employees of the Institute of Immunology, a producer of the L-Zagreb mumps vaccine strain.












