INTRODUCTION
Noroviruses (NoVs) are not only the leading causative agents of outbreaks of acute viral gastroenteritis worldwide in people of all ages, but also the second most common viral aetiological agents of severe childhood gastroenteritis after rotavirus [Reference Patel1, Reference Glass, Parashar and Estes2]. Patel and co-workers [Reference Patel1] estimated that in children aged <5 years residing in resource-rich countries, NoVs cause ~900 000 episodes of gastroenteritis necessitating a clinic visit, compared with resource-poor countries in which NoVs may cause more than one million hospitalizations and up to 200 000 deaths each year.
A high incidence of NoVs (20%) in acute gastroenteritis (AGE) affecting young children was originally reported from Finland when stool specimens collected for rotavirus studies in connection with a rotavirus vaccine efficacy trial in 1993–1995 were also examined for human caliciviruses [Reference Pang, Joensuu and Vesikari3]. While many of the community-acquired NoV cases were mild, some were severe, and NoVs also accounted for about 10% of AGE cases seen in hospital [Reference Pang4]. In the present study, we examined the incidence and genotypes of NoVs in community-acquired AGE affecting young children during 1993–2007, a period when large numbers of infants and young children participated in several rotavirus vaccine trials in Finland.
Most NoVs infecting humans belong to genogroups GI and GII. GI genogroup is further subdivided into at least eight genotypes and GII genogroup into 17 genotypes [Reference Zheng5]. Of the two main genogroups, GII is much more common worldwide [Reference Atmar and Estes6, Reference Kroneman7]. Genotype GII.4 NoVs have predominated since the mid-1990s in the USA, Europe and Oceania, causing 70–80% of all NoV outbreaks in communities, nursing homes, schools, hospitals and cruise ships associated with contaminated food or water [Reference Kroneman8, Reference Zheng9]. GII.4 has also commonly been detected in endemic infections of children worldwide [Reference Bucardo10–Reference Cheng14]. Of the other genotypes, recombinant strain GIIb especially has also been frequently reported in paediatric NoV infections [Reference Lindell11–Reference Sdiri-Loulizi13, Reference Bruggink and Marshall15].
In the past, NoV genotyping was performed solely on the RNA-dependent RNA polymerase (RdRp) region of open reading frame (ORF) 1 of the single-stranded, positive-sense NoV RNA genome [Reference Katayama16]. Later studies showed better segregation of the different strains into their respective genotypes by phylogenetic analysis of nucleotide sequences within the capsid region of ORF2 [Reference Vinje, Hamidjaja and Sobsey17]. However, genotyping based solely on the capsid sequence would miss the naturally occurring recombinant NoVs which cluster into two distinct groups of NoV strains when regions RdRp and capsid are subjected to phylogenetic analysis [Reference Bull18, Reference Bull, Tanaka and White19]. In this study, we genotyped NoVs according to polymerase region as well as to the capsid region.
METHODS
Clinical specimens
In total, 4727 faecal specimens were collected from AGE cases in children up to age 3 years participating in seven rotavirus vaccine efficacy trials in several different regions in Finland from 1993 to 2007 [Reference Pang, Joensuu and Vesikari3, Reference Vesikari20–Reference Vesikari23]. The vaccine study protocols and consent forms had been approved by the appropriate ethics committees and patients or legal guardians volunteered for the study after having given informed consent. Stool specimens were collected from children receiving either placebo or rotavirus vaccine whenever an episode of AGE occurred. AGE was typically defined as ⩾3 looser than normal stools and/or vomiting within any day. Specimens collected within 14 days of the first sample from the same patient were considered to be duplicate samples and excluded from the analysis. The numbers of stool specimens from each year and the genotyping methods applied are described in detail in Table 1. Stool specimens were stored at −20°C until tested.
Table 1. Study specimens analysed and the methods used for RNA extraction and NoV genotyping 1993–2007
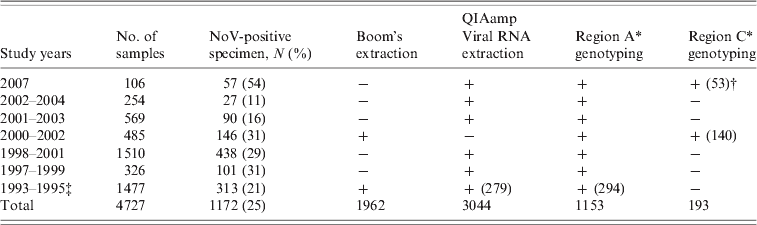
−, Not tested; +, tested.
* See Methods section and Figure 1.
† If not all specimens were tested the number in parentheses indicates the number of specimens tested.
‡ Results published in [Reference Pang4].
RNA extraction
For viral RNA extraction 10% (w/v) stool suspensions were made in phosphate-buffered saline (PBS). Extractions were performed using the QIAamp® Viral RNA Mini kit (Qiagen, Germany) according to the manufacturer's instructions. In some cases (Table 1), RNA was extracted by binding to silica particles in the presence of guanidine thiocyanate as described earlier by Boom et al. [Reference Boom24]. Aliquots of RNA extracts were stored at −70°C until reverse transcriptase (RT)–PCR testing.
RNA polymerase (region A) RT–PCR
For the specimens collected between 1994 and 1995 the detection of NoVs was performed using primer mixture sets including reverse primer NVp110 and forward primers NI, NVp69 and Np36 (Fig. 1) [Reference Le Guyader25]. These primers detect both genogroup I and II NoVs as well as sapovirus, and amplify 397 bp, 151 bp and 116 bp long PCR products from the RNA-dependent RNA polymerase (RdRp) region. RT–PCR reactions were performed as described previously [Reference Pang, Joensuu and Vesikari3].
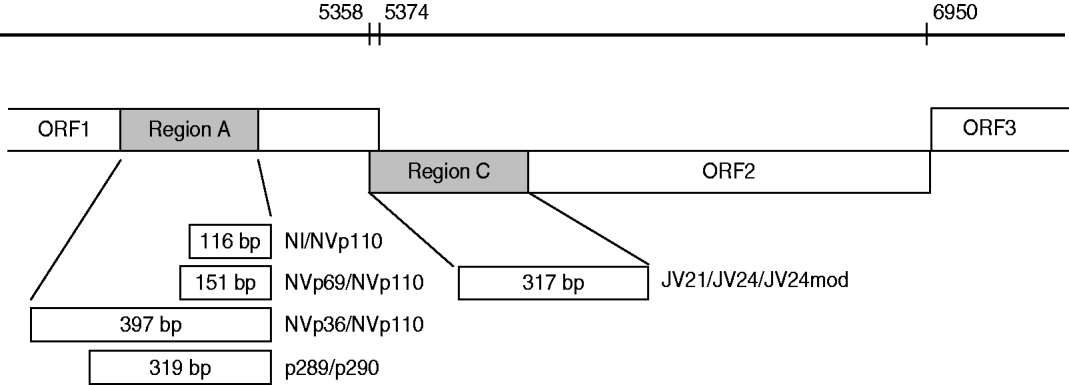
Fig. 1. Schematic presentation of norovirus genomic regions A and C (grey) [Reference Vinje, Hamidjaja and Sobsey17], and RT–PCR primers (NI/NVp69/NVp36/NVp110, p289H,I,IUB/p290H,I,J,K,IUB and JV21/JV24/JV24mod, described in the Methods section) used for reverse transcriptase (RT)–PCR and genotyping. Numbers on the bar refer to the nucleotide positions in Norwalk GI.1 virus genome (GenBank accession no. M87661). RT–PCR amplicon sizes in base pairs are noted in the boxes.
For the specimens collected between 1997 and 2007, as well as for 279 samples from 1994 to 1995 (to confirm genotype findings done by the RT–PCR method described above), RNA polymerase region A detection was performed by the RT–PCR method reported by Jiang and co-workers [Reference Jiang26], modified by Farkas and colleagues [Reference Farkas27]. The primer mixture p289H, I/p290H,I,J,K was used with additional primers p289IUB (reverse: 5′-GATTACTCCARGTGGGAYTCMAC-3′) and p290IUB (forward: 5′-TGACRATKTMATCATCMCCRTA-3′) to improve detection of all genotypes. The RT reaction was performed at 42°C for 60 min with 2·5 μl RNA added to the RT mixture (total volume 50 μl) containing 22·9 μl sterile water, 1×GeneAmp PCR buffer (Applied Biosystems, USA), 1·5 mm GeneAmp MgCl2 (Applied Biosystems), 400 μm dNTPs each, 16 ng/μl p289H,I,IUB reverse primer mixture, 10 U RNasin® (Promega, USA) and 70 U M-MLV Reverse Transcriptase RNase H− enzyme (Promega). Fifty microlitres of PCR reaction mixture consisting of 26·6 μl sterile water, 2 U GoTaq DNA polymerase (Promega), 1×GoTaq Green buffer (Promega), 0·5 mm MgCl2 (Promega), and a mixture of p290H,I,J,K,IUB forward primers (24 ng/μl) was added to the RT reaction. The 40-cycle PCR was run in a GeneAmp PCR system 9700 or Thermal Cycler 2720 (Applied Biosystems) with the following conditions: primary denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 42°C for 1 min 30 s, extension at 72°C for 1 min and final extension at 72°C for 10 min. The PCR amplicons were analysed by agarose gel electrophoresis to confirm the correct size of the product. This RT–PCR method for the detection of RdRp of caliciviruses simultaneously detected both NoVs and sapoviruses and amplified a 319-bp amplicon for NoVs and a 331-bp amplicon for sapoviruses. Positive PCR products were stored at −20°C for sequencing.
Capsid (region C) RT–PCR
To confirm the genotyping by region A, RT–PCR typing targeted at region C (Fig. 1) from the beginning of the capsid region in ORF2 of NoVs was studied for 193 specimens collected between 2000–2002 and 2007 (Table 1). Five microlitres of RNA was first reverse-transcribed as described by Pang et al. [Reference Pang, Preiksaitis and Lee28], except that the reaction contained 1× first-strand buffer (Invitrogen, USA) and the final concentration of dNTPs was 375 μm each. Synthesized cDNA was stored at −20°C unless used immediately for PCR reaction. A 317-bp fragment was amplified with primers JV21 (reverse), JV24 (forward) [Reference Buesa29] and an additional forward primer JV24mod (5′-GTGAATGAAGATGGCGTCGA-3′) for GII genotypes (Fig. 1). The PCR reaction was performed with 5 μl cDNA added to PCR mixture (total volume 50 μl) consisting of 22·5 μl sterile water, 2·5 U GoTaq DNA polymerase (Promega), 1× GoTaq Green buffer (Promega), 1 mm MgCl2 (Promega), 200 μm dNTP (Promega) and 4 ng/μl mixture of each of the above primers. The PCR run conditions were identical to those described above for RNA polymerase RT–PCR except that annealing temperature was 49°C.
Sequencing
All the positive PCR products were excised from the electrophoresis gel, purified with QIAquick® Gel Extraction kit (Qiagen) according to the manufacturer's instructions and sequenced using the Big Dye® Terminator v. 1.1 Cycle Sequencing kit (Applied Biosystems). The primers used for sequencing were identical to those used in the RT–PCRs. Ethanol precipitation purified sequencing PCR products were analysed using an automated sequencer ABI PRISM™ 310 Genetic Analyser (Applied Biosystems).
Sequence analysis and genotyping
Sequences obtained by the ABI PRISM 310 Genetic Analyser were aligned, verified and edited with the program Sequencher™ 4.8 software (Gene Codes Corp., USA). Virus confirmation and genotyping was done using the Food-borne Viruses in Europe network (FBVE) NoV genotyping tool (http://www.rivm.nl/bnwww) and NCBI Blast® programs (http://www.ncbi.nlm.nih.gov/BLAST/, nucleotide blast). In addition, RdRp region A and capsid region C (Fig. 1) sequences of the recombinant strains were re-analysed with the new NoV genotyping tool of FBVE (http://www.rivm.nl; National Institute of Public Health and the Environment, The Netherlands) to confirm the genotyping.
RESULTS
A total of 4727 faecal specimens from AGE cases were studied and 1172 (25%) cases were found to be associated with NoVs, as determined by genotyping according to region A sequences (Table 1). NoV genotypes during the period 1994–2007 by month are shown in Figure 2. The study period covered ten seasons with each season starting in July and ending in June. Of the 1149 genotyped NoV strains, 96% were genogroup GII. GII.4 was the most common genotype throughout the study period (46%), but it appeared in varying proportions in the NoV epidemic seasons (Fig. 2, Table 2). GII.4 was already present in community-acquired cases in 1993–1994 with a 40% share, but dominated in the seasons 1998–1999, 2002–2003 and 2006–2007 (Table 2, values in bold face).
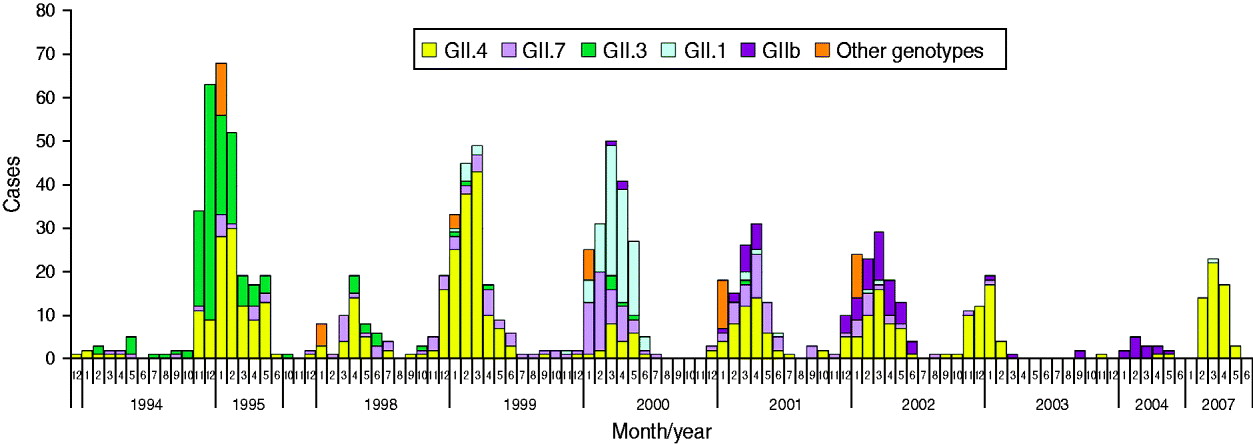
Fig. 2. Norovirus (NoV) seasonality and genotype distribution in Finnish children aged <3 years throughout the study period 1993–2007. From July 1995 to September 1997 and from June 2004 to January 2007 no specimens were collected.
Table 2. Most commonly found NoV genotypes in sporadic cases of acute gastroenteritis in Finnish children 1993–2007. From July 1995 to September 1997 and from June 2004 to January 2007 there were no specimens available
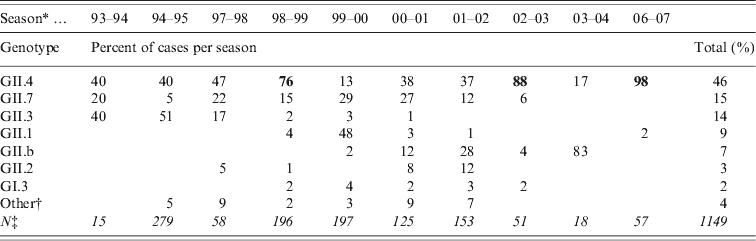
Bold values indicate peak years of GII.4 incidence.
* Season is a time period from July to June.
† Other genotypes detected (GI.6, GI.4, GII.9, GI.2, GIIU, GIId and undetermined).
‡ Total number of NoV-positive specimens (in italics).
Additional NoV genotypes detected are listed in Table 2. Other genotypes included: GI.6, GI.4, GII.9, GI.2, GIIU, GIId which accounted for <1% each (Fig. 2, Table 2). During the seasons 1999–2000 and 2003–2004, which followed the GII.4 peak seasons, the shares of GII.4 were low (13% and 17%, respectively), and genotypes GII.1, GII.7 and GII.b dominated. In the early study years GII.4 was circulating with GII.3 (1994–1998) and with GII.7 (1994–2002), followed by GII.1 (1999–2000) and GII.b in the later seasons (2000–2007). Overall, of the total ten study seasons GII.4 predominated in seven seasons, while other genotypes predominated only in one season each (Fig. 2, Table 2).
The second most common genotype throughout the 14 study years was GII.7, which appeared in multiple seasons but never dominated (Fig. 2, Table 2). Genotype GII.3 circulated largely in 1994–1995 (40–50%) and 1997–1998 (17%) but was not detected after 2001. Genotype GII.1 was detected only rarely in other seasons, but predominated in 2000. The first recombinant GII.b strains (GII.b–GII.2) appeared in March 2000. GII.b reached a high prevalence in 2001–2002 and was the dominating genotype in 2004. The most common GI strain GI.3 occurred during 1998–2003 with very low prevalence of 1–4%. Only a few AGE cases with other GI strains (GI.6, GI.4, GI.2) were occasionally detected (data not shown). In 1994–2002 there were more than eight different genotypes circulating among the study children, but later, in 2002–2007, the number of different genotypes was at maximum three per season (Table 2).
To confirm whether genotyping by capsid region C would identify identical genotypes as determined by polymerase region A, some of the specimens (years 2000–2002 and 2007, n=193) were genotyped for both of these regions (Fig. 1, Table 1). Identical genotyping results were obtained for both regions in all cases, except for the recombinant strains. Altogether 53 recombinant strains were found in 193 double genotyped NoVs in 2001–2002. No recombinant strains were found in 2007, when GII.4 was detected almost exclusively (Tables 1 and 3). RNA polymerase genotype GII.b occurred most commonly with capsid genotype GII.3 (41 cases, 77%) and rarely with GII.1 (three cases, 6%). Polymerase type GII.7 occurred with capsid genotype GII.6 (six cases, 11%) and GII.14 (one case, 2%) (Table 3).
Table 3. Polymerase (region A) and capsid (region C) genotype combinations of recombinant strains detected in Finnish children in 2001–2002

—, Undetermined.
DISCUSSION
Rotavirus vaccination studies conducted in Finland with follow-up AGE specimens offered a unique opportunity to study extensively the prevalence of NoVs in community-acquired AGE in infants and young children. Due to genetic variation in NoVs, primer selection for RT–PCR assays has been challenging. It is possible that in early study seasons from 1993 to 1995 NoV-positive specimens were missed when we used polymerase region primers including NVp110 [Reference Le Guyader25] which were shown to fail to detect some NoV strains [Reference Vinje30]. However, the large number of samples allowed us to draw certain conclusions. The present study showed that in children aged <3 years, NoVs were responsible for about one quarter of AGE cases in the community.
Overall, GII.4 was the most prevalent genotype in young children during the study period 1994–2007, followed by GII.7, GII.3, GII.1 and GII.b. In European outbreaks of AGE in all ages 2001–2006 the most common genotypes were GII.4, GII.7, GII.2 and GII.b [Reference Kroneman8]. In European outbreaks GII.7 peaked in 2005–2006 [Reference Kroneman8] and in Finnish outbreaks in 2001 and 2002 [Reference Maunula and Von Bonsdorff31]. However, in children we had already found GII.7 in earlier seasons (1997–1998 and 1999–2001). Consistent with the observations from outbreaks in Finland and other European countries [Reference Kroneman8, Reference Lindell11, Reference Maunula and Von Bonsdorff31] in winter 2001, GII.b was the third most common genotype, and its relative role increased until winter 2004, when it was the predominant type (83%). In contrast to our observation, in a Canadian study [Reference Lee32] no correlation was found between the NoV strains in the outbreaks and in sporadic childhood AGE, but the follow-up period was only a little over 1 year.
In our study, genogroup GI strains were uncommon and, therefore, appeared to be less common in sporadic cases in children than in outbreaks. In Finnish outbreaks in 1998–2002 GI strains were detected in 13%, but we detected GI in <6% [Reference Maunula and Von Bonsdorff31]. On the other hand, both in the outbreaks and in our study, GI.3 was the most common GI genotype [Reference Maunula and Von Bonsdorff31]. In general, GI strains occur throughout the year rather than seasonally, more often in water-mediated rather than person-to-person-mediated outbreaks, and only rarely in sporadic cases of NoV infections [Reference Zheng9, Reference Lysen33–Reference Bon35].
The total proportion of GII.4 NoV in young children was 46% for the years 1994–2007 and 52% for 2001–2007. Genotype GII.4 has also been detected as the predominant NoV genotype in paediatric sporadic AGE cases reported elsewhere [Reference Lindell11, Reference Sdiri-Loulizi13, Reference Cheng14, Reference Medici36]. The peaks of genotype GII.4 in winters 2002–2003 and 2006–2007 detected in Finland, occurred at the same time as peaks of GII.4 outbreaks elsewhere in Europe and the USA [Reference Kroneman8, Reference Zheng9]. In 1998–1999 GII.4 was also seen in Finnish outbreaks and this winter was also reported as a GII.4 outbreak peak year in Germany [Reference Maunula and Von Bonsdorff31, Reference Lopman37]. Furthermore, from 2006 to 2008 GII.4 was found in almost 90% of children hospitalized for NoV AGE (Räsänen et al., unpublished observations). An increase in NoV activity in outbreaks was seen in Finland and many other European countries in 2002 due to the emergence of a new GII.4 variant 2002 [Reference Kroneman8, Reference Maunula and Von Bonsdorff31, Reference Lopman37]. GII.4 variants 2006a and 2006b were responsible for large NoV outbreaks in Finland in winter 2007 [Reference Kanerva38]. Analyses of genetic variants of GII.4 in Finnish children for the peak years are ongoing and will determine which subtypes were responsible for these peaks.
Naturally occurring recombination events are common in NoVs and the most common recombination site is the ORF1–ORF2 junction localized upstream of the capsid gene [Reference Bull18, Reference Bull, Tanaka and White19]. We found 77% of the recombinants in our study consisted of a combination of GII.b–GII.3, which was also the most common recombinant strain elsewhere and emerged recently as a causative agent for many outbreaks in Europe, Australia, and Asia [Reference Bruggink and Marshall15, Reference Bull, Tanaka and White19, Reference Lysen33]. Genotype GII.b was the most common genotype in Finnish outbreaks in 2001 but was not detected in Finland before January 2001 [Reference Maunula, Miettinen and von Bonsdorff34] nor elsewhere in Europe before August 2000 [Reference Bon35, Reference Ambert-Balay39]. In the present study six GII.b cases had already been detected in March–April 2000. This raised a new possibility regarding the origin and transmission of the recombinant GII.b strains, which were previously suspected to be in contaminated shellfish in August 2000 [Reference Ambert-Balay39]. Children with sporadic gastroenteritis could serve as a reservoir for emerging epidemic NoV strains as some NoV strains appeared in children prior to emerging as epidemic outbreak strains [Reference Sdiri-Loulizi13, Reference Medici36]. To the best of our knowledge, we report for the first time the recombination of GII-7 polymerase with GII.14 capsid. However, the recombinant cases we described should be verified by sequencing over the ORF1–ORF2 junction region to exclude the possibility of mixed infections of different genotypes.
In conclusion, similar to findings worldwide, GII.4 was found to be the predominant NoV genotype in endemic gastroenteritis in infants and young children in Finland, at least since 1994. Recombinant genotype GII.b was detected with increasing incidence from March 2000 onwards. The shifts in NoV genotypes may be associated with an increase in the clinical significance of NoV AGE in young children overall, and warrant more in-depth studies on GII.4 variants.
ACKNOWLEDGEMENTS
Dr Shang-Qin Zeng and the technicians at the laboratory of the Vaccine Research Center at the University of Tampere Medical School are gratefully acknowledged for their excellent technical assistance during the study.
DECLARATION OF INTEREST
None.









