INTRODUCTION
Intestinal amoebiasis is caused by the protozoan Entamoeba (E.) histolytica, a non-flagellated amoeboid protozoan parasite. E. histolytica is an invasive pathogen commonly acquired in the developing world. However, most humans infected with E. histolytica are asymptomatic [Reference Benetton1]. When clinical symptoms develop, they are usually limited to the gastrointestinal tract. The symptoms of abdominal pain, tenderness and diarrhoea may mimic manifestation of irritable bowel syndrome (IBS) or inflammatory bowel disease. E. histolytica has the ability to lyse host cells and cause tissue destruction while it also induces both cellular and humoral immune responses in extraintestinal disease [Reference Ackers and Mirelman2].
The genus Entamoeba, including species E. histolytica, E. dispar, E. moshkovskii, E. poleki, E. coli and E. hartmanni may colonize the human intestinal lumen. E. histolytica is known to cause intestinal and extraintestinal disease while other species are regarded as commensal organisms that cause no intestinal disease. Faecal carriage of E. dispar is more common than E. histolytica. It is known that even in areas where invasive amoebiasis is common E. dispar is the more prevalent species [Reference Petri, Singh, Guerrant, Walker and Weller3]. Mixed infection with E. histolytica, E. dispar and/or E. moshkovskii have been reported [Reference Ali4]. The demonstration of cysts or trophozoites in the stool suggests an intestinal amoebic infection, but microscopy cannot differentiate between E. histolytica and E. dispar or E. moshkovskii.
Amoebiasis may form part of the differential diagnosis of IBS, especially in patients with acute exacerbations of IBS symptoms. Early studies implicated amoebic dysentery in the development of IBS in British soldiers returning from Egypt at the end of the Second World War [Reference McMillan5, Reference Stewart6]. However, in later years, several studies refuted this assertion and exposure to E. histolytica was thought not to predispose patients to IBS and all patients spontaneously eradicated the organism [Reference Chaudhary and Truelove7–Reference Sinha9]. In the current study, the prevalence of E. histolytica, E. dispar and E. moshkovskii was determined in patients who presented with IBS, i.e. chronic diarrhoea associated with abdominal pain and or discomfort.
MATERIALS AND METHODS
Patients
A total of 318 stool samples were examined between June 2008 and December 2009. They were obtained from 161 patients with chronic intermittent diarrhoea for the last 3 months associated with abdominal discomfort or pain mimicking IBS and ⩾3 loose stool per day, and 157 healthy controls with normal bowel habits who volunteered stool samples. Patients with IBS symptoms attended the gastroenterology clinic at the Aga Khan University, Karachi. Their mean age was 41±15 (range 16–83) years with a male:female ratio of 112:49. These patients underwent history, physical examination, complete blood count, serum creatinine, electrolytes, stool microscopy, culture and polymerase chain reaction (PCR) for E. histolytica, E. dispar and E. moshkovskii. Presence of chronic diseases, e.g. celiac disease, thyroid dysfunction and chronic pancreatitis were ruled out by tissue transglutaminase antibody IgA and IgG, thyroid-stimulating hormone and serum amylase level. In the control group, there were 157 healthy individuals without previous history of diarrhoea. They were comprised of medical and paramedical staff members who volunteered for a history, physical examination and a stool sample. Their age and sex closely matched the patient group with diarrhoea. Controls were all local residents of Karachi and randomly selected. They were excluded if they had a history of diarrhoea, foreign travel or were exposed to risk factors for infection during the last 6 weeks; they were known not to suffer from any comorbid illnesses that could contribute to chronic infection. The study was approved by the institutional ethics review committee of the Aga Khan University. All stool specimens were processed by microscopy and culture for Entamoeba and the presence of other parasites such as Blastocystis hominis, Giardia lamblia, E. coli, E. hartmanni, etc. was noted. DNA used for PCR was extracted from both unpreserved stool specimen and Entamoeba culture to ensure that the organism which grew was the same as that seen by the microscope or detected by PCR. The Entamoeba genus-specific primers were first used with extracted DNA to detect Entamoeba spp. which was followed by PCR with species-specific primers to detect E. histolytica, E. dispar and E. moshkovskii.
A microbiological investigation was also performed to detect Salmonella spp., Shigella sonnei, Campylobacter jejuni, Clostridium difficile and Vibrio cholerae. However, a viral screen was not performed on stool specimens.
Microscopy of stool smear
Stool sample microscopy consisted of examining ~2 mg emulsified faeces with one drop of physiological saline and Lugol's iodine covered with a coverslip on two separate glass slides. Later an ethanol-fixed faecal smear was stained with modified trichrome stain. These preparations were examined under both low (×10) and high (×40–100) power. The diameter of the cysts was measured and the nuclei were counted. A 4-nucleated cyst having a diameter ranging from 10 to 16 μm was identified as an Entamoeba cyst.
Culture of stool specimen
Cultures were performed for Entamoeba spp. in Robinson's medium shortly after collection as previously described [Reference Robinson10]. The cultures were incubated at 37°C and examined after 2–3 days. The culture was deemed negative if no growth was observed after 5 days. The sediment was examined as described above.
Extraction of genomic DNA
Stool DNA was extracted by using Stool DNA Extraction kit (Qiagen, USA) according to the manufacturer's protocol. Extracted DNA was stored at −20°C until PCR was performed for Entamoeba spp.
PCR for Entamoeba spp
The primers used were as previously described (Table 1) [Reference Khairnar and Khairnar11]. These primers detected nucleotide sequences of 16S-like ribosomal RNA gene (16S rRNA) of E. histolytica, E. dispar and E. moshkovskii. For genus-specific and species-specific PCR, the reaction volume of 25 μl comprised 2·5 μl of 10× PCR buffer (Promega, USA), 2·0 μl of 25 mm MgCl2 (Promega), 0·75 μl deoxyribonucleotide triphosphate mix (10 mm each dNTP, Promega), 0·5 μl (5 IU/μl) Taq polymerase (Promega), 0·25 μm primers (IDT) and 2·0 μl template DNA. Amplification was performed in a PerkinElmer 9700 thermal cycler with both positive and negative controls. Positive controls consisted of DNA that has been positive twice for the same Entamoeba species while distilled water was used as the negative control. The PCR conditions consisted of one cycle denaturing at 94°C for 5 min, 35 cycles including annealing at 55°C for 1 min, extension at 72°C for 1 min, denaturing at 94°C for 1 min, and an additional cycle with a 5-min chain elongation at 72°C (PCR System 9700, PerkinElmer, USA). The PCR products and molecular markers were electrophoresed in 2% agarose gel with Tris-acetate-EDTA electropheresis buffer. The size markers were 100-bp ladders (Promega). The PCR amplification for each primer pair was repeated at least three times. Bands were visualized by the imaging system (Gel Doc 2000, Gel Documentation System, Bio-Rad, UK) after being stained with ethidium bromide.
Statistical method
Results were expressed as mean±standard deviation for continuous variables (e.g. age) and number (percentage) for categorical data (e.g. gender, stool culture, diarrhoea, etc.). Univariate analysis was performed by using the independent-sample t test, Pearson's χ2 test and Fisher's exact test where appropriate. The kappa (κ) test was used to compare methods. A P value <0·05 was considered statistically significant. All P values were two-sided. Statistical interpretation of data was performed by using the computerized software program SPSS version 16.0 (SPSS Inc. USA).
RESULTS
Stool microscopy with modified trichrome stain for Entamoeba cyst was positive in 36% (115/318) of samples. Entamoeba culture was positive in 38% (121/318). Entamoeba genus PCR was positive in 39% (125/318) with E. dispar positive in 19% (59/318), E. moshkovskii in 13% (42/318) and E. histolytica in 7% (21/318), while it was non-typable in 3% (8/318) of samples. Co-infection with B. hominis was seen in 12% (38/318) of samples by stool culture. These comprised of 20% (32/161) of patients and 4% (6/157) in the control group. These patients with B. hominis co-infection were excluded and the final analysis was performed in 280 patients, 129 (46%) with diarrhoea and 151 (54%) controls. Endolimax nana cysts were noted in six (2%) samples which were equally present in both groups. Bacterial cultures were negative in these patients for Salmonella spp., Sh. sonnei, C. jejuni, Cl. difficile and V. cholerae.
Association of symptoms with Entamoeba spp
Patients with diarrhoea had E. histolytica infection (9%, 11/129) compared to 1% (2/151) of controls (P=0·004) while 19% (24/129) of patients had E. moshkovskii infection compared to 4% (6/151) of controls (P<0·001) (Table 2). E. dispar was present in 19% (24/129) of patients with diarrhoea compared to 27% (40/151) of controls (P=0·679).0
Table 2. Details of patients included in the study
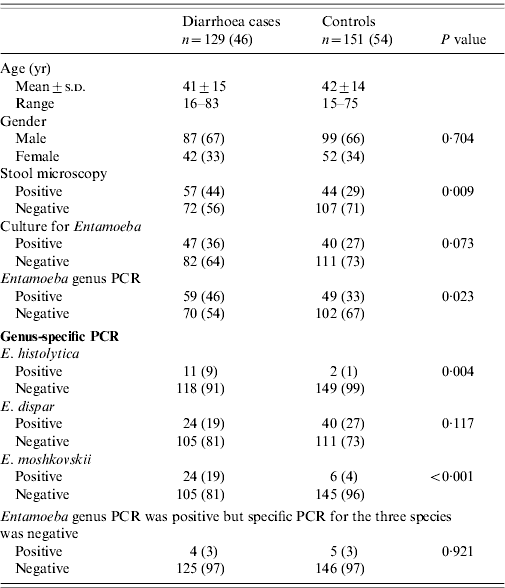
Univariate analysis was performed by using the independent-sample t test, Pearson's χ2 test and Fisher's exact test were also used whenever appropriate. A P value <0·05 was considered statistically significant.
Values are number and percentage: n (%).
Diagnostic yield of various tests used for the diagnosis of Entamoeba spp
Stool microscopy with staining demonstrated Entamoeba cysts in 44% (57/129) of patients with diarrhoea compared to (44/151) of controls (P=0·009). Entamoeba culture was positive in 36% (47/129) of patients compared to 27% (40/151) of controls (P=0·073). One patient had mixed infection with E. moshkovskii and E. dispar. PCR for Entamoeba genus was positive in 46% (59/129) of patients compared to 33% (49/151) of controls (P=0·023). E. histolytica was positive in 9% (11/129) of patients compared to 1% (2/151) of controls (P=0·008) (Table 2). E. moshkovskii was positive in 19% (24/129) of patients compared to 4% (6/151) of controls (P<0·001) (Table 2).
Correlation between stool microscopy and PCR for Entamoeba spp
Entamoeba cysts were observed by stool microscopy in 44% (57/129) of patients with diarrhoea while PCR for E. histolytica was positive in 9% (11/129) (κ=0·14, P<0·001), E. dispar in 19% (24/129) (κ=0·21, P=0·004) and E. moshkovskii in 19% (24/129) (κ=0·21, P=0·004), respectively, compared to when Entamoeba cysts were negative (56%, 72/129).
DISCUSSION
Clinical amoebiasis may mimic functional bowel disease when it has a subacute onset with symptoms of mild diarrhoea and abdominal pain [12]. E. dispar and E. moshkovskii infections and 90% of E. histolytica infections were reported as being asymptomatic [12]. This study showed that in patients with diarrhoea and abdominal pain, Entamoeba cysts were commonly seen on stool microscopy. Diarrhoea was associated with both E. histolytica and E. moshkovskii infection. These patients were treated with metronidazole and diloxanide combination following which they had symptomatic improvement with a clear repeat stool examination. In our study we did not use a concentration step such as formol-ether centrifugation which is known to increase the sensitivity of microscopy as the diarrhoeal stool samples were usually liquid and did not concentrate well. The direct stool smear technique which is quick and inexpensive was preferred for the observation of motile protozoan trophozoites. It is possible that by not using a concentration technique, we may have missed some parasites if their concentration was low or if much debris was present. We did not anticipate any difference in the sensitivity of stool testing based on its consistency between cases and controls, as most of our cases described their stool consistency as loose and not watery. Further, clinical details of the patient were not known at the time of stool microscopic examination. The yield of stool culture and Entamoeba genus PCR was better than microscopy. E. dispar species was present equally in both groups. Only two patients with diarrhoea had infection with two different types of Entamoeba spp. In these cases, E. moshkovskii was associated with E. histolytica or E. dispar. E. histolytica was significantly associated with diarrhoea and was found in only one in healthy control who did not have a history of diarrhoea (Table 2). In a few cases, we were unable to type the Entamoeba species which might represent E. coli and E. hartmanni that are common commensals in the intestinal tract of the humans.
In this study, E. moshkovskii was demonstrated as the second most common Entamoeba spp. E. dispar is described as a non-pathogen although it has been shown to be capable of producing variable focal intestinal lesions in animals and destroyed epithelial cell monolayers in vitro [Reference Chadee, Smith and Meerovitch13–Reference Espinosa-Cantellano15]. E. moshkovskii cysts are morphologically indistinguishable from those of E. histolytica and E. dispar. In a previous study, prevalence of E. moshkovskii infection of 21% was reported in children aged 2–5 years in Bangladesh [Reference Ali4]. E. dispar-infected children were twice as likely to be co-infected with E. moshkovskii (35%) compared to those with E. histolytica (18%) infection [Reference Ali4]. A study in India has also linked E. moshkovskii infection with dysentery [Reference Parija and Khairnar16]. Similarly, in a study from Australia that did not use a control group, the prevalence of E. moshkovskii infection was 61·8% in homosexual men and all these patients were symptomatic [Reference Fotedar17].
This is the first report to study the molecular typing of Entamoeba spp. in Pakistan and highlights the incidence of co-infection in our patients. It is important to recognize that routine stool microscopic examination techniques do not impart complete information to allow differentiatiation between the Entamoeba spp. Co-infection with Entamoeba spp. might be responsible for exacerbation of symptoms in some of the known IBS cases. The reporting of Entamoeba cysts by stool microscopy might not be suggestive of a benign course. There might be co-infection with more than one type of Entamoeba spp. that could not be identified on routine stool microscopic examination. It is possible that E. dispar in association with E. moshkovskii may contribute to the development of diarrhoea.
ACKNOWLEDGEMENTS
We are grateful to the staff of the Juma Research Laboratory for their help during the completion of this work.
DECLARATION OF INTEREST
None.







