Article contents
A versatile method for generating single DNA molecule patterns: Through the combination of directed capillary assembly and (micro/nano) contact printing
Published online by Cambridge University Press: 24 January 2011
Abstract
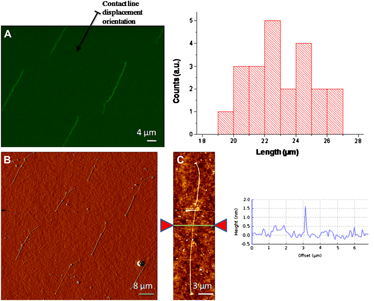
One of the challenges in the development of molecular scale devices is the integration of nano-objects or molecules onto desired locations on a surface. This integration comprises their accurate positioning, their alignment, and the preservation of their functionality. Here, we proved how capillary assembly in combination with soft lithography can be used to perform DNA molecular combing to generate chips of isolated DNA strands for genetic analysis and diagnosis. The assembly of DNA molecules is achieved on a topologically micropatterned polydimethylsiloxane stamp inducing almost simultaneously the trapping and stretching of single molecules. The DNA molecules are then transferred onto aminopropyltriethoxysilane-coated surfaces. In fact, this technique offers the possibility to tightly control the experimental parameters to direct the assembly process. This technique does not induce a selection in size of the objects, therefore it can handle complex solutions of long (tens of kbp) but also shorter (a few thousands of bp) molecules directly in solution to allow the construction of future one-dimensional nanoscale building templates.
- Type
- Reviews
- Information
- Journal of Materials Research , Volume 26 , Issue 2: Focus Issue: Self-Assembly and Directed Assembly of Advanced Materials , 28 January 2011 , pp. 336 - 346
- Copyright
- Copyright © Materials Research Society 2011
References
REFERENCES
- 14
- Cited by




