INTRODUCTION
Salmonellosis is the most frequent foodborne disease reported in France, often transmitted by foods of animal origin, especially raw eggs, egg products or poultry meats, causing more than 300 outbreaks per year [Reference Haeghebaert, Le Querrec, Gallay, Bouvet, Gomez and Vaillant1]. To address this public health problem, national surveillance networks to monitor Salmonellae of both human and non-human origins were set up. These networks are used to reveal trends in the evolution of different serotypes, monitor their antimicrobial susceptibility, detect outbreaks, and contribute to their investigation. Antimicrobial resistance monitoring has become more important since the appearance and dissemination of multidrug-resistant Salmonella Typhimurium phage type DT104, conferring resistance to ampicillin, streptomycin, sulphonamide, chloramphenicol and tetracycline (resistance-type ASSuCT). The genes encoding for these resistances are clustered on the chromosomal genomic island SGI1 and can be transferred clonally and also horizontally to other serotypes [Reference Cloeckaert and Schwarz2, Reference Threlfall3]. The increasing evolution of resistance-type ASSuCT has been reported in several countries since the 1990s [Reference Breuil, Brisabois, Casin, Armand-Lefevre, Fremy and Collatz4–Reference Murphy, McNamara, Hill and et7]. Besides this resistance type, other antimicrobial resistances, notably to quinolones, appeared initially in developing countries [Reference Hart and Kariuki8] and then worldwide [Reference Hakanen, Siitonen, Kotilainen and Huovinen9, Reference Threlfall, Ward and Rowe10]. Many reports suggest that the main cause of increasing antimicrobial resistance among Salmonella isolates from animals is the misuse and overuse of antimicrobials in the breeding sector, and that some of these strains may be transmitted through food to humans, thereby contributing to the increase of antimicrobial resistance among humans [Reference Aarestrup11–Reference van den Bogaard and Stobberingh13]. These high antimicrobial resistance levels in zoonotic Salmonella may contribute to an increase of morbidity in human infections because resistant bacteria may be more virulent and the treatment less effective [Reference Barza and Travers14, Reference Travers and Barza15].
The purpose of this descriptive study was to evaluate trends in antimicrobial resistance in the main Salmonella enterica serotypes of human and poultry sources over 8 years in France, by reviewing data collected annually in 1993, 1997 and 2000. The following five predominant serotypes were chosen for the study: Typhimurium, Enteritidis, Hadar, Heidelberg and Virchow. Observed trends are discussed with respect to data on veterinary antimicrobial consumption in France.
MATERIALS AND METHODS
Bacterial isolates
Surveillance of isolates from animal and food origin has been carried out by the French Agency for Food Safety (AFSSA) through the ‘Salmonella’ Network. Strains were classified in three main sectors: (i) animal health; meaning poultry, pig, cattle or other farm animal isolates and nearby environment isolates, representing more than 70% of the total collected isolates; (ii) food hygiene; meaning isolates from animal food products, feedstuffs and slaughter houses, for 25% of isolates; (iii) ecosystems; meaning water purification plant, harbour and sewage isolates corresponding to <5% of isolates. However, the third sector did not have a sufficiently large sample size and was not used in this analysis.
Strains were collected on the basis of voluntary participation of veterinary and food hygiene laboratories, located throughout the national area. All collected strains were sent with information concerning the origin, animal species, categories of food and regional localization of the samples. The samples were mainly collected within the framework of self-regulating systems based on legislation requirements or on random official sampling (eggs) and monitoring plans (poultry farming), and within the animal breeding environment.
The French Salmonella Reference Centre (CNRS) at the Pasteur Institute (Paris, France) has conducted the surveillance of human isolates by collecting specification sheets and isolates from public and private clinical laboratories of analysis for several decades. The strains have been isolated from clinical samples (blood or faeces), and sent to the CNRS on the basis of voluntary participation. Human strains were selected annually for susceptibility testing, in respect of the distribution of serotypes.
Serotyping and antimicrobial susceptibility testing
Serotype was determined by the Kauffmann–White method, using Bio-Rad (formerly Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) standard antisera and home-made antisera. This study reports the results observed for the five serotypes: Typhimurium, Enteritidis, Hadar, Virchow and Heidelberg, selected on the basis of their high frequency of isolation both in human and non-human origins (Table 1). The antimicrobial susceptibility was tested by standard disk diffusion on Mueller–Hinton agar plates according to the guidelines of the Comité de l'Antibiogramme de la Société Française de Microbiologie (CASFM).
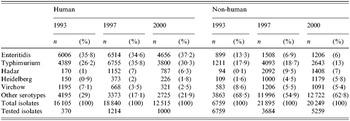
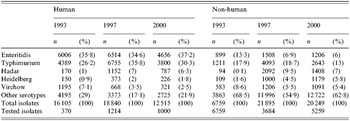
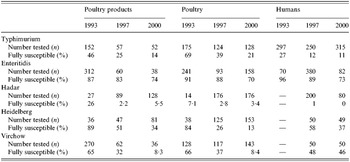
Table 1. Annual number of reported Salmonella isolates from human and non-human origins in five studied serotypes
Zone diameters were automatically read by the Osiris system (Bio-Rad) using the break-points defined by the guidelines of CASFM, and have been interpreted as predictive of treatment effectiveness classes: resistant (R), intermediate (I), susceptible (S). Strains classified as intermediate class were considered in this study as susceptible. E. coli ATCC 25 922 strain was used for quality control.
Ten antimicrobial drugs and two combinations of antimicrobials have been tested on all isolates as follows (disk content indicated in parentheses): nalidixic acid (30 μg), ampicillin (10 μg) for non-human isolates, amoxicillin (10 μg) for human isolates, amoxicillin-clavulanic acid (20 μg+10 μg), streptomycin (10 IU), trimethoprim–sulphamethoxazole (1·25 μg+23·75 μg), trimethoprim (5 μg), sulphonamide (200 μg), tetracycline (30 IU), chloramphenicol (30 μg), gentamycin (10 IU), cefotaxime (30 μg), ofloxacin (5 μg). These listed antibiomicrobials correspond to those recommended for the surveillance of antimicrobial susceptibility by Caprioli et al. [Reference Caprioli, Busani, Martel and Helmuth16] and by the European group (Office International des Epizooties). Sulphonamide was not tested in animal isolates in 1993. Concerning the ampicillin and amoxicillin disks tested, we considered that their cross-susceptibilities and cross-resistances were complete [Reference Fuchs, Barry, Pfaller, Hardy, McLaughlin and Gerlach17]. Table 2 gives the number of tested strains and fully susceptible strains for each studied serotype.
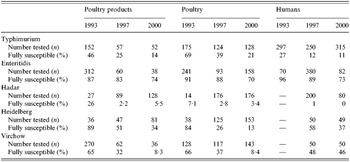
Table 2. Number of strains tested for each serotype and percentage of fully susceptible strains
Statistical analysis
Statistical analyses were performed using Epi-Info software (version 6.04b, CDC, Atlanta, GA, USA). We assessed the emergence of new R-types, possibly clustered in a gene, by comparing the observed rate of associated resistance to the predicted rate if these resistances were occurring independently, using the McNemar test. The χ2 trends test was used for evaluating evolution of resistance to nalidixic acid and ofloxacin over the 8 years. P values <0·05 were considered as significant.
RESULTS
Frequency of isolation and global rate of resistance (Tables 1 and 2)
S. Typhimurium was the second most common serotype found in human isolates and the most common in animal isolates. The proportion of human S. Typhimurium susceptible to all the antimicrobials tested decreased significantly over the 8 years (27% to 11%, P<10−5).
S. Enteritidis was the most frequent serotype in human isolates among the serotypes tested and its rank was second or third among non-human isolates. Since 1993, susceptible human, animal and food isolates have been decreasing; nevertheless it remained the most susceptible serotype among those studied.
S. Hadar constituted <1% of all isolates in 1993 but increased rapidly between 1993 and 1997 both in human and non-human isolates. There were already few susceptible human isolates in 1997 (1%) and they totally disappeared in 2000.
Isolation of the serotype S. Heidelberg increased strongly among non-human strains between 1993 and 2000. The proportion of susceptible strains in human isolates decreased between 1997 and 2000 from 58% down to 37%. Although levels of resistance were very low to all drugs in 1993, the proportion of susceptible strains has steadily declined. Strains isolated from animals had generally higher resistance rates than those from poultry products.
S. Virchow isolation decreased between 1993 and 2000 in both human and non-human sectors. It was the only serotype in which the proportion of susceptible strains was stable in human isolates (48% of all isolates in 1997 and 46% in 2000). The proportion of susceptible isolates from the poultry sector steadily decreased during the 1993–2000 period.
ASSuCT R-type and new R-types (Table 3)
The frequency of resistance type (R-type) ASSuCT strains greatly increased among Typhimurium isolates between 1993 and 2000 (P<10−3). The occurrence of this R-type was much higher than would be predicted by the independent occurrence of each of the antimicrobials separately. In 2000, 40% of human isolates and 45% of poultry sector isolates harbouring the ASSuCT R-type were also nalidixic acid-resistant. Moreover, 43% of trimethoprim-resistant human isolates also presented the ASSuCT R-type. However, the ASSuCT R-type appears not to have disseminated among the other serotypes.
Table 3. Percentages of ASSuCT R-type and other association of resistances among the five serotypes

Sulphonamide was not tested.
Even without any spread of R-type ASSuCT, the serotype Hadar has shown the most spectacular increase in resistance over the 8 years with the lowest rate of susceptible strains among the five serotypes. Resistances to ampicillin or amoxicillin, streptomycin and tetracycline, with or without the resistance to nalidixic acid often occurred together but they were not statistically associated in a particular R-type.
In Heidelberg, human strains showed approximately the same antimicrobial resistance as those from poultry products, with an increase in rates between 1997 and 2000. A novel R-type ASSuT, emerged strongly from 4% in 1997 to 54% in 2000 for animal isolates. This R-type was detected in 6% of human isolates in 1997 and represented 22% in 2000, commonly with additional resistance to trimethoprim. These R-type values were much higher than the predicted values based on the occurrence of each antimicrobial separately and confirmed the co-occurrence of these resistances were not independent.
Resistance to nalidixic acid and to ofloxacin (Table 4)
Strains of Typhimurium resistant to nalidixic acid were already present in 1993 and increased significantly between 1993 and 2000 for the three sectors.
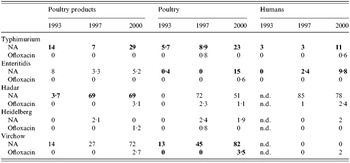
Table 4. Percentages of resistance of the five serotypes to nalidixic acid (NA) and ofloxacin
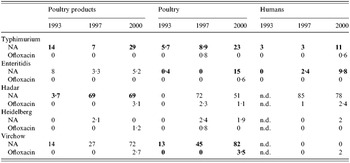
Significant increases among the three years detected by χ2 trends test appear in bold (P<0·05).
For Enteritidis, resistance rates to nalidixic acid increased significantly among poultry and human isolates between 1993 and 2000. In most cases, a single resistance was observed and concerned 89% of nalidixic acid-resistant strains for the poultry sector in 2000.
Among S. Hadar, resistance rates to nalidixic acid rose steeply between 1993 and 2000 in the animal and food poultry sectors. Seventy-six per cent of nalidixic acid-resistant strains in the poultry sector isolated in 1997 and 84% of those isolated in 2000 were found occurring together with the resistance to amoxicillin or ampicillin, streptomycin and tetracycline. Among human isolates, this pattern of resistance was observed for 78 and 69% of nalidixic acid-resistant strains in 1997 and 2000 respectively. Resistance to ofloxacin also appeared in the three sectors.
Over these 8 years, resistance rates of S. Virchow to nalidixic acid in poultry isolates greatly increased. Resistance to ofloxacin also appeared in 2000 among human and poultry isolates. The resistance to nalidixic acid was found as a single resistance or in combination with other resistances. Among the nalidixic acid-resistant strains, occurrences of single resistance were: 44% in 1993, 63% in 1997, 56% in 2000 in the poultry sector, and 42% in 1997, and 83% in 2000 strains from humans.
DISCUSSION
Our study revealed an increase in antimicrobial resistance rates for all Salmonella isolates from poultry, poultry product and human origin over this 8-year period, except for S. Virchow human isolates. Although the R-type ASSuCT did not disseminate from Typhimurium to other serotypes, we noted the emergence of a new R-type ASSuT in Salmonella serotype Heidelberg, and a worrying increase of resistance to nalidixic acid and even to ofloxacin among the five serotypes studied, especially among Hadar and Virchow.
Considering that the collection of isolates was based on voluntary participation, there was a selection bias, amplified by the application of regulatory rules in some animal production sectors (e.g. poultry sector). Therefore, the results cannot be considered as completely representative and exhaustive.
Our results may have been overestimated because some information concerning the context of sample collecting was not available. Indeed, in cases of human disease, an antimicrobial treatment could have been administered before collecting stool samples and may have selected resistant strains. Nevertheless, the geographical distribution and the number of samples were large enough to reveal trends in antimicrobial resistance during this 8-year period.
Since Salmonella infection is a zoonotic disease, antimicrobial resistance of strains from animals and humans have been frequently linked to antimicrobial use in agriculture [Reference Breuil, Brisabois, Casin, Armand-Lefevre, Fremy and Collatz4, Reference Murphy, McNamara, Hill and et7, Reference Aarestrup11–Reference van den Bogaard and Stobberingh13, Reference Franklin, Acar and Anthony18–Reference White, Zhao and Sudler24]. For this reason, monitoring of antimicrobial resistance among Salmonella from food-animals and humans on the one hand, and of antimicrobial use in agriculture, on the other, has been initiated recently in France [Reference Martel, Tardy, Brisabois, Lailler, Coudert and Chaslus-Dancla25, Reference Martel, Tardy, Sanders and Boisseau26] and worldwide (WHO, DANMAP, Euro-surveillance). The comparison between the evolution of antimicrobial resistance and antibiotic use in agriculture is complicated, first because of the lack of statistics in this sector, and second due to the presence of multiple interaction factors. Nevertheless, our observations can be evaluated based on the first study of veterinary antimicrobials sold in France in 1999 [G. Moulin, personal communication, 1999]. During that year, tetracycline, aminoglycoside, ampicillin or amoxicillin and sulphonamides composed 83% of the antibiotics used as veterinary drugs in animals. In this study, the highest rates of resistance were observed among these antimicrobial families, as shown by the association of resistance to ampicillin, streptomycin and teracycline in serotype Hadar, or the emergence of the R-type ASSuT in serotype Heidelberg. In contrast, no resistance to chloramphenicol was observed among serotype Heidelberg, although it acquired the R-type ASSuT. One hypothesis for this observation is that Heidelberg was rare before 1994, the year chloramphenicol was banned. Therefore, Heidelberg strains have not been exposed to a selective pressure by this antibiotic. Comparing R-type ASSuT occurrences observed with the predictive values, suggests that genes encoding this R-type may be clustered on the same genetic element. Molecular analysis should be conducted in order to assess the organization of genetic elements associated with this new R-type.
Similarly, the highest levels of ASSuCT resistance were observed among S. Typhimurium, both in poultry and in human isolates. This could be explained by the clonal spread of Typhimurium DT104, linked to the well-documented pattern of antimicrobial resistance ASSuCT [Reference Cloeckaert and Schwarz2–Reference Breuil, Brisabois, Casin, Armand-Lefevre, Fremy and Collatz4, Reference Murphy, McNamara, Hill and et7, Reference Poppe, Smart, Khakhria, Johnson, Spika and Prescott22, Reference Rabsch, Tschape and Baumler23]. In our study, frequency of ASSuCT R-type strains remained stable for the serotype Typhimurium between 1997 and 2000 but were 10-fold higher than the predictive values given by the occurrence of each antimicrobial drug independently. Moreover, no evidence of large horizontal dissemination of this R-type was observed even if some previous results showed the presence of the genomic island structure SGI1 in other serotypes such as Agona, Paratyphi B and Albany [27].
Furthermore, besides regulation on the use of specific antimicrobials, policies to eradicate infected flocks may also have played a role. For instance, since 1998 poultry flocks infested by S. Typhimurium and S. Enteritidis have been eradicated in accordance with the CEE zoonosis directive (no. 92/117). The frequency of isolation of S. Enteritidis is stable among human strains, and low in non-human strains, probably due to these legislative requirements.
We also observed the emergence of nalidixic acid resistance among human and poultry isolates in S. Typhimurium. Resistance to quinolones was studied in 2001 by Cloeckaert and colleagues regarding the emergence of Typhimurium DT104 after the introduction of enrofloxacin in veterinary medicine in 1993 [Reference Cloeckaert and Schwarz2, Reference Cloeckaert, Sidi Boumedine, Flaujac, Imberechts, D'Hooghe and Chaslus Dancla27, Reference Cloeckaert and Chaslus-Dancla28]. In 1999 in France, quinolones represented 1·5% of the total of quantities of antimicrobial drugs sold, which is relatively low, but the available data cannot allow us to analyse a correlation between consumption and resistance. Oral formulations of quinolones and fluoroquinolones have been authorized for the treatment of E. coli infection in poultry. When a prophylactic or curative treatment is given to the whole flock or herd, via mixing with feed or water, some resistant strains can emerge and spread through animals not taking antibiotics at a bactericidal concentration [Reference Bager and Helmuth19, Reference Johnston29].
Cases of treatment failure of ceftriaxone and decreased susceptibility to ciprofloxacin in human isolates have been observed [Reference Fey, Safranek and Rupp21, Reference Threlfall, Skinner, Graham, Ward and Smith30, Reference Walker, Lawson and Lindsay31]. Furthermore, some studies demonstrate the presence of subpopulations with resistant or decreased susceptibility to fluoroquinolone among the nalidixic acid-resistant isolates, which might influence the outcome of fluoroquinolone treatment [Reference Hakanen, Kotilainen, Jalava, Siitonen and Huovinen32]. In 1999, Threlfall drew our attention to the increase of isolated strains with decreased susceptibility to ciprofloxacin among strains belonging to serotypes Virchow and Hadar in England [Reference Threlfall, Ward, Skinner and Graham33]. In our study, these two serotypes had the highest rates of resistance to nalidixic acid. The most striking increase of resistance concerned the serotype Hadar with the frequent association of nalidixic acid resistance to resistance to amoxicillin, streptomycin and tetracycline. In Finland, between 1995 and 1997, quinolone-resistant strains of S. Hadar harboured a resistance to streptomycin, tetracycline and sometimes to ampicillin or amoxicillin [Reference Hakanen, Siitonen, Kotilainen and Huovinen9]. Our results are also in accord with Cruchaga et al., who observed that the ASTNal resistance pattern with additional cephalotin resistance was the most frequent R-type [Reference Cruchaga, Echeita, Aladuena, Garcia-Pena, Frias and Usera5]. With regard to our results, it should be noted that resistance to ofloxacin emerged in 2000 among S. Hadar human strains.
The results of our analyses show some trends in the evolution of antimicrobial resistances in Salmonella strains in France and led us to detect some new R-type emergence in the five most-studied serotypes. The resistance monitoring system must be improved and should be compared with the distribution of veterinary antimicrobials. Such improved surveillance will be necessary to analyse the mechanisms and modes of dissemination, and public health impact of both the emerging resistances and the pathogens themselves.
ACKNOWLEDGEMENTS
The authors thank V. Vaillant of the Institut de Veille Sanitaire (InVS) for expert assistance. Part of this work has been supported by InVS. We gratefully acknowledge Mrs Jayne Ireland and Mrs Claire Evans (AFSSA) for greatly improving the English grammar and style of the manuscript.
DECLARATION OF INTEREST
None.