Article contents
Associative overdominance caused by linked detrimental mutations*
Published online by Cambridge University Press: 14 April 2009
Summary
Associative overdominance due to linked detrimental mutations was investigated using the method of moment equations based on diffusion models. The expectation of the apparent selective value at the marker (neutral) locus has been evaluated. Assume two linked loci, at one of which the steady flux equilibrium is reached under constant mutational input of deleterious mutations (with rate v) having disadvantages hs in heterozygote and s in homozygotes. At another locus, the neutral alleles are segregating with frequencies near 0·5. Let Ne be the effective size of the population and c be the recombination fraction between the two loci. Then the coefficient of associative overdominance at the neutral locus can be obtained by taking the expectation with respect to chromosome frequencies at steady flux equilibrium. It becomes approximately
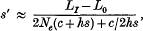
where (LI−L0) is the inbreeding depression caused by deleterious mutations under complete inbreeding, and Nehs ≫ l and hs ≫ v are assumed. More generally, if the inbreeding depression of a chromosome segment with a length of recombination fraction C is (LI−L0) then s′ at the neutral marker at the edge of the segment is
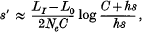
where hs is the average heterozygote disadvantage of detrimentals.
The significance of the associative overdominance is discussed in relation to actual observations. It is proposed that the most of the observed heterozygote superiority including inversion chromosomes of Drosophila, isozyme alleles in Avena and ABO blood group genes in man could be explained by the associated detrimentals.
- Type
- Research Article
- Information
- Copyright
- Copyright © Cambridge University Press 1971
References
REFERENCES
- 160
- Cited by




