Since their first report in Korea in 1992, vancomycin-resistant enterococci (VRE) have become endemic as an important nosocomial pathogen [Reference Lee1]. In earlier periods, most VRE infections in Korea occurred in large tertiary-care or university hospitals [Reference Lee1] and since then, most clinical research on VRE colonization and infection in Korea has focused on isolates from tertiary-care hospitals, but there is a lack of molecular epidemiological studies on VRE isolates from primary- or secondary-care hospitals.
VRE is an important concern because infections with these organisms are difficult to treat not only in clinical practice, but they can also spread within and between hospitals and further to different regions or countries. Molecular epidemiological studies are necessary to clarify the genetic relatedness and molecular evolution of such clones and thereby inform patterns of spread and consequently measures for infection control. This study aims to examine the evolutionary relationships of VRE isolates from non-tertiary and tertiary-care hospitals in Korea by a comparative analysis of the antimicrobial and molecular characteristics of isolates.
We studied a total of 62 clinical vancomycin-resistant Enterococcus faecium (VREF) isolates from diverse geographical areas, including an island, around Korea from October 2010 and April 2011. Thirty-six isolates were collected from 20 primary- or secondary-care hospitals including small medical institutions, medium-sized general hospitals, and geriatric care hospitals (0–544 beds). For comparison, 26 isolates were collected from three different university hospitals (1087–2093 beds). The latter are teaching tertiary-care hospitals with organ transplantation services, cancer centres, emergency services, intensive care units, and various medical specialities in Seoul and Suwon, Korea. VREF were identified by conventional biochemical reactions with the Vitek identification system (bioMérieux Inc., USA) and the API Strep kit system (bioMérieux Inc.). We analysed the minimum inhibitory concentrations (MICs) of vancomycin and teicoplanin, resistance genotype, transposon (Tn) 1546 structures, multilocus sequence typing (MLST), and a dendrogram of DNA fingerprinting created by automated repetitive extragenic palindromic PCR (rep-PCR) in all VREF isolates. VREF strain BM4147 was used as a control. Antimicrobial susceptibility testing of vancomycin and teicoplanin for VREF was performed by the E-test according to Clinical and Laboratory Standards Institute (CLSI) recommendations [2]. DNA of VREF was extracted from agar grown cultures with the Qiagen DNeasy kit (Qiagen GmbH, Germany) according to the manufacturer's instructions, and multiplex PCR was performed to investigate the presence of vanA, vanB, vanC1, and vanC2/C3 genes as published previously [Reference Dutka-Malen, Evers and Courvalin3]. For structural analysis of Tn1546, PCR amplification of overlapping internal regions of Tn1546 was performed as described previously [Reference Lee4, Reference Simonsen5]. The purified PCR products were directly sequenced on an ABI Prism 3100 machine (Applied Biosystems, USA) and were analysed using DNASIS for Windows v. 2.6 (Hitachi Solutions America Ltd, USA). MLST was performed on VREF isolates according to Homan et al. [Reference Homan6]. The allele number for each gene was assigned according to the E. faecium MLST database (http://www.efaecium.mlst.net). Automated rep-PCR analysis was performed for bacterial strain typing and determination of genetic relatedness of the isolates as previously described [Reference Healy7]. DNA for rep-PCR analysis was extracted with the UltraClean™ Microbial DNA Isolation kit (MoBio Laboratories Inc., USA) according to the manufacturer's instructions. Data were analysed with the web-based DiversiLab® Analysis software version 3.4 (bioMérieux, USA), the Pearson correlation coefficient was calculated to determine distance matrices, and the unweighted pair-group method with arithmetic mean (UPGMA) was used to create dendrograms.
As with most VREF isolates found in Korea and worldwide [Reference Lee1], all of the isolates studied here harboured the vanA resistance gene and additionally were negative for vanB, vanC1, and vanC2/C3 genes. The vanA gene typically corresponds to the VanA phenotype which is characterized by high-level resistance to both vancomycin and teicoplanin, whereas the vanB gene is responsible for the VanB phenotype (resistance to vancomycin but not to teicoplanin) [Reference Dutka-Malen, Evers and Courvalin3, Reference Lauderdale8]. However, the van genotype in this study was not completely consistent with the phenotype as shown in earlier reports [Reference Lee4, Reference Lauderdale8]. The MICs of vancomycin and teicoplanin for VREF isolates from the non-tertiary-care hospitals ranged from 48 to > 256 μg/ml, and from 2 to >256 μg/ml, respectively. All tertiary-care isolates showed vancomycin MICs >256 μg/ml and teicoplanin MICs from 4 to 96 μg/ml. Fifteen (41·7%) of 36 isolates from non-tertiary-care hospitals and 19/26 (73·1%) from tertiary-care centres exhibited the VanB phenotype: vancomycin MICs of 48 to >256 μg/ml and teicoplanin MICs of 2–16 μg/ml (Table 1). We used the breakpoint of teicoplanin for defining the VanB phenotype according to Lauderdale et al. [Reference Lauderdale8]. The vanA genotype-VanB phenotype VREF isolates were more frequently identified in tertiary-care than non-tertiary-care hospitals (P < 0·05, χ 2 test). The molecular basis of the vanA genotype-VanB phenotype discrepancy has yet to be identified, but it has been suggested that impairment of accessory proteins VanY and VanZ, genetic rearrangement including deletion of both vanY and vanZ genes following insertion of IS1216V, or mutations in the vanS regulatory gene may be responsible for the loss of teicoplanin resistance [Reference Lee4, Reference Simonsen5, Reference Lauderdale8].
Table 1. Characteristics of 62 vancomycin-resistant E. faecium isolates from non-tertiary-care and tertiary-care hospitals in Korea

MLST, Multilocus sequence typing; MIC, minimum inhibitory concentration; VM, Vancomycin; TEI, teicoplanin; ST, sequence type; GB, Gyeongbuk; GG, Gyeonggi; GN, Gyeongnam; GW, Gangwon; JB, Jeonbuk; JJ, Jeju island; SE, Seoul.
In this study, Tn1546 structure analysis showed that none of the isolates was identical to the prototype (E. faecium BM4147) and no mutations were identified in the central vanS regulatory gene regions. Insertion sequence (IS) 1542 or IS1216V-IS1542 was inserted at nucleotide position 3932 in the orf2-vanR intergenic region, and IS1216V was located within or downstream of vanX in all isolates, and complete or partial deletion of nucleotides adjacent to the IS sites was evident in all. The isolates were classified into three main types according to Tn1546 structural differences from the prototype (Fig. 1). Type I isolates (n = 5, 8·1%) were characterized by an IS1542 insertion in the orf2-vanR intergenic region and IS1216V insertion in the vanX-vanY intergenic region. Type II isolates (n = 39, 62·9%) had IS1542 and IS1216V inserted at the left end of the Tn1546 with a deletion of orf1 and/or orf2 regions as well as IS1216V insertion in the vanX-vanY intergenic region, and in type III isolates (n = 18, 29%) the left and right ends of Tn1546 were totally deleted and accompanied with IS insertion. Fifteen (83·3%) of the type III group representing the deletion of vanY and vanZ, showed the VanB phenotype, while 15 isolates (44·1%) of the VanB phenotype were Tn1546 type III. The complete deletion of both vanY and vanZ genes followed by an insertion of IS1216V may be one of factors responsible for the impaired resistance to teicoplanin.
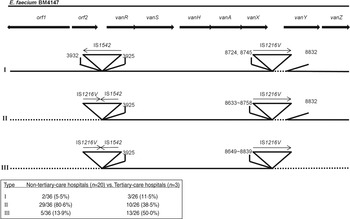
Fig. 1. Genetic maps of Tn1546 types of 62 vancomycin-resistant E. faecium isolates from Korean hospitals. The positions of genes, open reading frames (orf1 and orf2) and the direction of transcription are marked by arrows at the top. The inverted triangles with vertical lines represent insertion sequence (IS) elements. Deletions are indicated by dotted lines. The positions of the first nucleotide upstream and the first nucleotide downstream from the IS insertion sites are depicted.
Tn1546 structural analyses, in addition to traditional methods like pulsed-field gel electrophoresis typing, have been used to investigate dissemination mechanisms of VRE [Reference Simonsen5]. The genetic rearrangements of Tn1546 reported so far include point mutations, insertion of IS elements, and deletions of the left (orf1 side) ends and/or the right (vanZ side) ends of the transposon [Reference Lee4, Reference Lauderdale8, Reference Schouten9]. We found that regardless of the hospital size, Tn1546 exhibited IS1542 and IS1216V in the genomes of all 62 isolates from diverse geographical areas of Korea (Fig. 1). Either IS1542 or IS1216V-IS1542 was inserted into the orf2-vanR intergenic region at nucleotide position 3932 in all isolates which is consistent with other studies [Reference Lee4, Reference Schouten9]. However, different integration sites of IS1216V in the vanX-vanY intergenic region indicated that the strains did not originate from clonal spread. This suggests that horizontal transfer of Tn1546 with IS1542 and IS1216V may have already occurred throughout Korea. IS1542 appears to be restricted to clinical and poultry VRE isolates from Europe, China, and Korea, while IS1216V of vanA elements is ubiquitous [Reference Lee1, Reference Schouten9–Reference Park11].
Internal size variations due to the presence of IS elements in Tn1546-like elements have been described previously [Reference Park11]. In this study, IS insertions were accompanied by complete or partial deletion of nucleotides adjacent to the insertion site. Park et al. reported that the rearrangement of Tn1546 such as integration of IS elements and deletions of nucleotides was associated with the evolution of the vanA gene cluster, and these elements may modify its transferability [Reference Park11]. Tn1546 sequences tended to be shortened as time passed, especially at the left or the right end of the transposon, and their transferability increased which suggests that the truncated sequences may play a role in the rapid dissemination of VRE [Reference Lee1, Reference Park11]. There was no significant difference in the incidence of Tn1546 type I (5·5%, 11·5%) between the non-tertiary and tertiary-care hospitals [P > 0·05, comparison of two rates by MedCalc version 12.7 (MedCalc Software, Belgium)]. The incidence of type II (80·6%, 38·5%) was significantly higher in the non-tertiary hospitals, whereas type III (13·9%, 50·0%) was more frequent in the tertiary-care hospitals (P < 0·05). As Tn1546 type increased, the number of VREF from the tertiary-care hospitals increased [P < 0·05, linear by linear association by SPSS software v. 17·0 (SPSS Inc., USA)]. Tn1546 type III was defined as the shortest type reducing their lengths by deleting sequences adjacent to IS elements. In tertiary-care hospitals, VRE have been exposed to adverse conditions such as intensive use of antibiotics or a variety of newly developed antibiotics over long periods, and we assumed that such genetic rearrangement most likely occurred due to an evolutionary adaptation of VRE to the hospital environment.
MLST is generally acknowledged to be an appropriate technique to establish an unambiguous international database of E. faecium genetic lineages in different laboratories or countries [Reference Homan6] and has been increasingly used to investigate the molecular evolution of VRE in regional and long-term global epidemiological studies [Reference Lee1, Reference Homan6]. We identified eight different sequence types (STs) of clonal complex 17 (CC17) in the 62 VREF isolates investigated (Table 1). ST17 (25·8%), the predicted founder of CC17, was predominant followed by ST192 (22·6%), ST78 (17·7%), ST18 (12·9%), ST262 (6·5%), ST202 (4·8%), ST203 (4·8%) and ST205 (4·8%). The regional distribution of these sequence types are shown in Supplementary Figure S1. CC17 represents the globally dispersed nosocomially adapted clonal lineage [Reference Lee1] and organisms in this complex are typically ampicillin- and quinolone-resistant and are enriched for putative virulence factors that may assist with hospital adaptation and spread [Reference Lee1]. CC17 is therefore an example of evolutionary processes that have improved the relative fitness of bacteria in the hospital environment. It has shown rapid spread and subsequent acquisition of vanA or vanB genes [Reference Lee1]. ST17 (2/26, 7·7%) was the least common of the isolates from tertiary-care hospitals, but was the most common type (14/36, 38·9%) from the non-tertiary-care hospitals (Table 1). This most likely reflects a shift in the predominant sequence type in tertiary-care hospitals. ST192, ST78, ST18, ST202, ST203 and ST205 are single or double locus variants of ST17 while ST262 is a single locus variant of ST18. ST262, which has been reported in VREF isolates from Europe and Russia [Reference Bourdon12, Reference Brilliantova13], was detected only in isolates from the non-tertiary-care hospitals in diverse geographical areas (Table 1). There was no association between Tn1546 type and MLST type. The dendrogram created with the automated rep-PCR results showed no clonal relatedness between the intra- and inter-hospital VREF isolates (data not shown). Taken together, these findings strongly suggest that vanA with IS1542 and IS1216V have spread horizontally throughout Korea from tertiary-care hospitals to non-tertiary-care hospitals.
A limitation of this study is that the numbers of isolates investigated may be insufficient to chart unequivocally the evolutionary relationships between VREF from non-tertiary and tertiary-care hospitals and further investigations are needed to elucidate the mechanisms of transmission and molecular evolution of these organisms in tertiary-care hospitals.
In summary, all VREF isolates harboured vanA with IS1542 and IS1216V integrated into Tn1546. IS1542 was inserted at the same position in the orf2-vanR intergenic region of all the isolates. Although all isolates belonged to hospital-adapted CC17, a shift in the dominant sequence type in tertiary-care hospitals may have occurred. VREF from the latter hospitals tended to exhibit a VanB phenotype and shortened Tn1546 lengths by the deletion of sequences adjacent to IS elements. We assume that the genetic rearrangements of VREF isolates from tertiary-care hospitals occurred during the continuing process of evolutionary adaptation.
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268813003543.
DECLARATION OF INTEREST
None.






