Increasing cases of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of Mycobacterium tuberculosis (MTB) continue to emerge, particularly from Asia and Africa [1]. In the USA, the majority of reported MTB cases have occurred in foreign-born persons [2]. MDR strains are resistant to rifampin and isoniazid, while XDR strains are resistant to rifampin and isoniazid plus one fluoroquinolone and at least one of the injectable drugs amikacin, kanamycin, or capreomycin. However, some MDR strains have additional drug resistance, i.e. pyrazinamide and/or streptomycin that might decrease treatment efficiency. Strains that become more highly resistant may, if unrecognized, become XDR strains by acquired drug resistance while on inadequate therapy.
MDR and XDR strains are typically characterized using drug susceptibility testing (DST), a procedure that is time-consuming (from weeks to months), involves propagation of MTB, and is technically challenging. An Ion Torrent full-gene sequencing approach (Ion Torrent, Life Technologies, USA [Reference Daum3]) was recently developed for rapid characterization of MTB resistance mutations [Reference Daum3] that can be performed for clinical isolates within days.
In this case-study report, the developed full-gene length Ion Torrent sequencing approach was expanded to include additional resistance genes for characterizing MDR and XDR mutations from clinical isolates within 5 days. We describe antibiotic resistance profiles deduced by Ion Torrent sequencing from three unique clinical isolates (two MDR and one unique drug resistant) obtained from Burmese, Hmong, and Indian immigrants residing in the USA. Ion Torrent sequencing data was compared to phenotypic data obtained by DST using the Bactec MGIT 960 (BD Diagnostic Systems, USA). Between April 2009 and November 2011, sputum specimens were collected from three patients using standard methodology at the City of Milwaukee Health Department's (MHD) Tuberculosis Control Clinic (TBCC) of the Keenan Health Center (KHC, Milwaukee, WI, USA).
Case 1. A 21-year-old Burmese woman entered the USA in April 2009 after staying in a refugee camp in Thailand. Her father was treated for pan-sensitive active tuberculosis (TB), and her mother and two siblings had latent TB infection. The patient was considered an active case based upon a preliminary investigation (smear-negative but chest X-ray abnormal, QuantiFERON test-positive). Initial treatment was standard therapy for pan-sensitive active tuberculosis: rifampin, isoniazid, pyrazinamide, and ethambutol.
Case 2. A 22-year-old Hmong woman entered the USA in September 2009 after staying at a different refugee camp than case 1 in Thailand. Initial chest X-ray, sputum, and QuantiFERON tests were negative. In August 2011, the patient was negative by smear microscopy but exhibited an abnormal chest X-ray and was treated at TBCC. The patient was started on rifampin, pyrazinamide and levofloxacin when M. tuberculosis grew on sputum culture. This regimen was chosen because the patient was a close household contact of a person with isoniazid-resistant, pulmonary TB. Later, due to intolerable, but innocuous side-effects, levofloxacin was changed to moxifloxacin. Case 2 completed 6 months of therapy.
Case 3. A 32-year-old woman from India entered the USA in May 2011 on a student visa. The patient presented with a prior history of TB (pan-sensitive in 2009), and developed high fever and cough 6 months after entering the USA. She was treated at TBCC where a preliminary evaluation revealed smear-positive sputum and an abnormal chest X-ray with cavitary lung lesions. Initial treatment was standard therapy for pan-sensitive active TB: rifampin, isoniazid, pyrazinamide, and ethambutol.
Primary clinical specimens (sputum) were confirmed for M. tuberculosis at the MHD laboratory using smear microscopy, MTB primer-specific real-time PCR [Reference Desjardin4], and by culturing with the BACTEC 960 system (BD Diagnostic Systems). Phenotypic resistance was determined using the MGIT 960 broth system [Reference Alcaide5]. Isoniazid (0·2 and 1·0 μg/ml), rifampin (1·0 μg/ml), and pyrazinamide (100 μg/ml) were used for DST testing.
Clinical isolates were handled in a blind fashion using the H37Rv strain as an internal sequencing control throughout the study. In a BSL-3 laboratory, a standard loop of MTB colonies grown on solid Löwenstein–Jensen medium was first inactivated in PrimeStore Molecular Transport Medium (MTM; Longhorn Vaccines & Diagnostics, USA), and then shipped at ambient temperature for sequencing. Total DNA was purified from 200 μl aliquots of PrimeStore MTM aliquots containing inactivated culture using the PrimeExtract System according to the manufacturer's protocol (Longhorn Vaccines & Diagnostics).
PCR amplification using full-gene primers for rpoB, katG, pncA, gyrA, and rrs were performed as described previously [Reference Daum3]. Additional primers for full-gene amplification of rpsL (forward rpsL: 5′-CATGGCCGACAAACAGAAC-3′ and reverse rpsL: 5′-TTGACCAACGGACGCTTG-3′), and inhA (forward inhA: 5′-AGCAGGACGGCATCAAAT and reverse inhA: 5′-CTGACACAACACAAGGACG) were designed and used for streptomycin and isoniazid resistance, respectively. PCR amplicons from each clinical isolate were pooled, sheared, size-selected, and subjected to adaptor/barcoding ligation as described previously [Reference Daum3]. Ion Torrent sequencing was performed according to the manufacturer's instructions (Life Technologies, USA) using 314 V2 chips with 200-bp sequencing chemistry. Sequencing runs produced average read lengths of 107 bp, greater than 30 000 high-quality (AQ20) reads, and a 300 × average depth of coverage. Next-generation assembly, sequence alignments, and protein translations were performed using SeqMan NGen (V4) and LaserGene (V10) Core Suite (DNAStar, USA). Gene sequences for rpoB, KatG, inhA, pncA, rpsL, gyrA, and rrs for cases 1–3 (TB11-91, TB11-111, and TB11-153, respectively) were deposited in GenBank (accession numbers KC692347–KC692364).
Ion Torrent sequencing of full-length rpoB, katG, inhA, pncA, rpsL, gyrA, and rrs genes was compared to phenotypic results obtained using the BACTEC MGIT 960 System (Table 1). Ion Torrent sequencing showed 100% concordance with phenotypic DST through direct identification of amino-acid substitutions known to confer antibiotic resistance (Table 1). Summarized in Table 2 are amino-acid changes that confer resistance as well as other discovered polymorphisms that differ from the H37Rv control.
Table 1. Comparison of genotypic and phenotypic drug resistance* Ion Torrent sequencing† and drug sensitivity testing
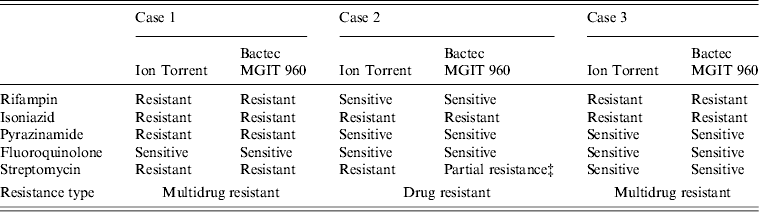
* Critical concentrations for indicated antibiotics are: rifampin (2·0 μg/ml); isoniazid (0·1 μg/ml and 0·4 μg/ml); pyrazinamide (100 μg/ml); fluoroquinolone (2 μg/ml); streptomycin (2·0 μg/ml and 6·0 μg/ml).
† Previously confirmed amino-acid substitutions giving rise to antibiotic resistance.
‡ Resistant at 2·0 μg/ml but sensitive at 6·0 μg/ml.
Table 2. Amino-acid substitutions in drug resistance genes from M. tuberculosis samples isolated from Burmese, Hmong, and Indian immigrants residing in the USA using Ion Torrent sequencing*
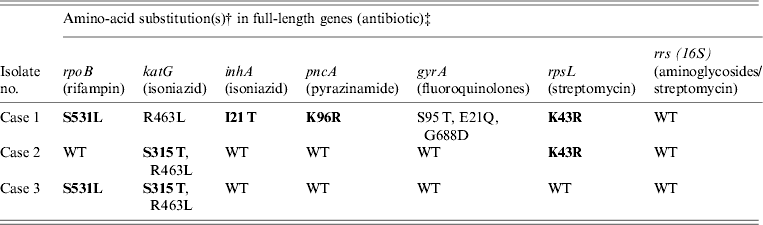
WT refers to the ‘wild type’ H37Rv reference strain.
Amino-acid changes in bold indicate a known resistance mutation.
* Compared to previously confirmed amino-acid substitutions known to confer antibiotic resistance.
† Compared to H37Rv wild-type reference strain.
‡ Gene sequences: case 1 (TB11-91), case 2 (TB11-111), and case 3 (TB11-153) were deposited in Genbank (accession numbers KC692347–KC692364).
As shown in Table 2, cases 1 and 3 contained a characteristic Ser531Leu mutation associated with rifampin resistance [Reference Zhang, Vilcheze and Jacobs6], while case 2 contained the wild-type residue. No other amino-acid changes were noted in the rpoB protein compared to H37Rv control.
All three cases were shown to be resistant to isoniazid at both low and high levels (0·1 μg/ml and 0·4 μg/ml) by phenotypic MGIT 960 testing (Table 1). Ion Torrent sequencing revealed that cases 2 and 3 exhibited the common Ser315Thr katG mutation known to confer between 75% and 94% of isoniazid resistance worldwide [Reference Mokrousov7]. All three strains also contained an Arg463Leu (R463L) change in katG that is not associated with isoniazid resistance but was recently observed within a subset of South African strains [Reference Daum3]. Case 1 did not contain any katG gene mutations, but exhibited a much rarer Iso21Thr (I21 T) substitution in the inhA gene that was previously correlated with isoniazid resistance [Reference Ho8].
Case 1 was shown by phenotypic DST to be resistant to pyrazinamide, while cases 2 and 3 were sensitive. Sequencing revealed a novel Lys96Arg (K96R) pncA substitution mutation in the case 1 isolate that had not been formerly reported (Table 2). Previous studies have characterized mutations at position 96 that include Glu (E), Gln (Q), Thr (T), or Asn (N) [Reference Lee, Lee and Jung9, Reference Lemaitre10] while a stop codon [Reference Mphahlele11] and frameshift mutation [Reference Denkin12] have also been reported. Case 1 is a highly resistant MDR strain and the additional PZA resistance may complicate treatment (Table 2).
Genetic characterization of the gyrA gene revealed that cases 2 and 3 exhibited a 100% sequence homology to H37Rv control sequence (Table 2). Case 1 was shown to contain three amino-acid substitutions (S95 T, E21Q, G688D) that differ compared to H37RV control (Table 2), but these mutations are not implemented in fluoroquinolone drug resistance and have been reported previously [Reference Daum3]. There were no mutations within the quinolone resistance determining region (QRDR), thus all three are sensitive to fluoroquinolone drugs according to Ion Torrent sequencing and DST (Tables 1 and 2).
Streptomycin resistance is mostly attributed to mutations in the 16S rrs and rpsL genes [Reference Zhang and Telenti13]. All three M. tuberculosis isolates exhibited 100% rrs nucleotide homology compared to H37Rv control, and are therefore sensitive to second-line aminoglycosides. However, cases 1 and 2 contained a Lys43Arg (K43R) substitution in the rpsL gene (Table 2). The rpsL gene which encodes the S12 ribosomal protein is associated with 42–59% of streptomycin-resistant strains, and the K43R mutation is known to cause resistance [Reference Zhang and Telenti13].
All three clinical isolates exhibited antibiotic resistance. Two were MDR (cases 1 and 3), and case 1 demonstrated additional resistance to streptomycin and pyrazinamide (Tables 1 and 2). Since case 2 was highly resistant to isoniazid (0·1 and 0·4 μg/ml) but sensitive to rifampin, rifampin-resistance detection alone may not be a good means for defining MDR and resistance to isoniazid [Reference Chang14]. All three isolates were phenotypically resistant to isoniazid; although, resistance was conferred from mutations residing on two separate genes (katG or inhA; Table 2). Case 1 containing the less common isoniazid-conferring (I21 T) mutation would have been missed by the HAIN line probe assay (LPA), a widely used drug-resistance genotyping assay [Reference Hillemann, Rüsch-Gerdes and Richter15].
A few commercial nucleic-acid-based assays for M. tuberculosis drug resistance have been developed and approved for use by the World Health Organization (WHO). The Hain Lifescience GenoType MTBDRplus and MTBDRsl LPAs (Hain Lifescience, Germany) are widely used for MDR and XDR detection [Reference Hillemann, Rüsch-Gerdes and Richter15]. The testing methodology is relatively simplistic and can screen smear-positive sputum specimens for a limited set of drug-resistant gene mutations in 1–2 days, but not without several drawbacks. There are less common and yet-to-be discovered mutations that confer resistance which are not covered by the limited space available on the LPA test strip. In this study, the case 1 isolate contained a less common isoniazid-conferring (I21 T) mutation that would not have been detected by Hain LPA (Table 2). Additionally, the LPA will not directly detect an amino-acid change even within a known site where the substitution represents an uncommon residue. In these cases, the absence of a wild-type band is the sole indicator for resistance, but the type of amino-acid substitution remains unknown. Furthermore, an undefined mutation indicated by the absences of a wild-type band in LPA may not be phenotypically resistant, and mixed strain populations with amino acids that are both wild-type and mutant may give ambiguous results. Characterizing the type of amino-acid substitution may be particularly important for phenotypic DST comparison to understand geographical variability in TB resistance. In contrast to LPA, our Ion Torrent full-gene sequencing approach allows for the potential discovery of new mutations that may be associated with resistance. However, genotypic discovery will require subsequent phenotypic minimal inhibitory concentration (MIC) studies to confirm resistance.
Ion Torrent sequencing could be used for routine MTB drug resistance characterization, and offers several advantages of traditional Sanger sequencing. In contrast to Sanger, the Ion Torrent does not utilize laser detection or light-sensitive chemistries that require fluorescent-labelled moieties, and has a small working footprint. The Ion OneTouch™ 2 System (Life Technologies, USA) for emulsification PCR, recovery and enrichment has improved the overall workflow, consistency, and timeliness in library preparation. An additional advantage of Ion Torrent sequencing over conventional Sanger sequencing is depth of coverage and characterization of mixed-strain populations. Furthermore, preliminary studies with Ion Torrent have shown full-length pncA gene characterization directly from primary samples collected and shipped in PrimeStore MTM with the limiting factor being the initial DNA concentration present in sputum samples [Reference Daum16]. Future studies are planned to develop an amplification and library preparation from primary sputum samples.
Drug-resistant TB is emerging in many parts of the world; however, characterization of these strains remains incomplete. In order to understand resistance mechanisms, guide clinical treatment, and develop public health interventions geographically diverse population studies are needed to identify phenotypic/genotypic resistance to first- and second-line drugs. Use of inappropriate or intermittent TB therapeutic measures may lead to the emergence of new resistant strains and challenge effective therapy. Therefore, timely gene sequencing will allow identification of new TB mutations, optimize effective treatment regimens, and ultimately support new drug development.
Political unrest, war, poverty, and famine will continue to lead to migration of refugees and immigrants from TB-endemic areas into the USA. Effective clinical treatment and public health control of TB will require identification of the emerging new mutations and drug-resistant TB strains. The next resistant TB strain may be just one plane journey away.
ACKNOWLEDGEMENTS
We acknowledge assistance from Irmine Reitl, Richard Wojciechowski, and David Schmid of the Milwaukee Health Department's Disease Control and Environmental Health (DCEH) unit for TB case investigations. We value the technical assistance from David Griswold, Bradley Krause, and Jose Navidad at the MHD laboratory who carried out the TB culturing and real-time PCR amplifications. We appreciate the Wisconsin State Laboratory of Hygiene and the CDC Mycobacteriology Laboratory Branch for additional confirmatory testing and second-line drug analysis. We also thank Julie Becker of the MHDL for her critical review of the manuscript.
DECLARATION OF INTEREST
Dr Luke T. Daum is the Chief Scientific Officer at Longhorn Vaccines & Diagnostics (San Antonio, TX, USA). His research interests include molecular epidemiology of infectious diseases including M. tuberculosis and influenza virus. Dr Gerald W. Fischer is the CEO and President of Longhorn Vaccines & Diagnostics. His work focuses on diagnostics and vaccines for infectious diseases.






