1. Introduction
The selfish DNA hypothesis proposes that the abundance of transposable elements (TEs) in natural populations of their hosts is the consequence of a balance between the proliferation of elements by transposition and natural selection acting to remove insertions from the population (Doolittle & Sapienza, Reference Doolittle and Sapienza1980; Orgel & Crick, Reference Orgel and Crick1980). Accordingly, any factor that modulates the strength of selection or rates of transposition should have an important effect on the distribution and dynamics of element insertions. At the genomic level, the impact of differences in the efficacy of selection on the patterning of TEs can be seen in the genomes of many organisms, and elements have been found to accumulate differentially among chromosomal regions in association with recombination rate differences (Charlesworth & Langley, Reference Charlesworth and Langley1989; Duret et al., Reference Duret, Marais and Biémont2000; Boissinot et al., Reference Boissinot, Entezam and Furano2001; Bartolomé et al., Reference Bartolomé, Maside and Charlesworth2002; Rizzon et al., Reference Rizzon, Marais, Gouy and Biémont2002, Reference Rizzon, Martin, Marais, Duret, Segalat and Biémont2003), base composition patterns (Lander et al., Reference Lander, Linton, Birren, Nusbaum, Zody, Baldwin, Devon, Dewar, Doyle and Fitzhugh2001) and gene density (Medstrand et al., Reference Medstrand, van de Lagemaat and Mager2002).
At the population level, a potentially important factor for TE dynamics is the host breeding system (Hickey, Reference Hickey1982; Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1995; Wright & Schoen, Reference Wright and Schoen1999; Morgan, Reference Morgan2001). When populations do not mate randomly, elements can be affected in a number of ways with different expected consequences. Self-fertilization can lead to element accumulation, because inbreeding reduces the effective population size experienced by the host, and smaller populations tend to accumulate more elements (Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1983; Brookfield & Badge, Reference Brookfield and Badge1997; Wright & Schoen, Reference Wright and Schoen1999; Morgan, Reference Morgan2001). In an inbreeding population, insertions will also tend to become homozygous: If the deleterious effects of TEs are caused by ectopic recombination between dispersed, heterozygous insertions (Montgomery et al., Reference Montgomery, Huang, Langley and Judd1991), selection may be more effective in outcrossing populations. But selfing can also lead to the containment of TEs, and if individual insertions are harmful because they disrupt genes (Finnegan, Reference Finnegan, Lindsley and Zimm1992), inbreeding should enhance the efficacy of natural selection against TEs because of higher homozygosity (Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1995). In addition, the complete stochastic loss of elements in small, self-fertilizing populations is also possible (Wright & Schoen, Reference Wright and Schoen1999; Morgan, Reference Morgan2001). A final effect of selfing is that lower levels of genetic exchange in inbred hosts might influence the evolution of transposition rates themselves, leading to conditions favouring TEs that reduce their own activity levels (Charlesworth & Langley, Reference Charlesworth and Langley1986; Charlesworth & Charlesworth, Reference Charlesworth and Charlesworth1995).
Comparisons of the population frequencies of TEs in natural populations have been performed within and between many animal species, including fruitflies (Charlesworth & Langley, Reference Charlesworth and Langley1989; Petrov et al., Reference Petrov, Aminetzrach, Davis, Bensasson and Hirsh2003; Bartolomé & Maside, Reference Bartolomé and Maside2004; Franchini et al., Reference Franchini, Ganko and McDonald2004), mosquitoes (O'Brochta et al., Reference O'Brochta, Subramanian, Orsetti, Peckham, Nolan, Arensburger, Atkinson and Charlwood2006; Boulesteix et al., Reference Boulesteix, Simard, Antonio-Nkondjio, Awono-Ambene, Fontenille and Biémont2007), midges (Zampicinini et al., Reference Zampicinini, Blinov, Cervella, Guryev and Sella2004), several fish species (Takasaki et al., Reference Takasaki, Amaki, Hamada, Park and Okada1997; Duvernell & Turner, Reference Duvernell and Turner1999; Neafsey et al., Reference Neafsey, Blumenstiel and Hartl2004) and humans (Batzer & Deininger, Reference Batzer and Deininger2002; Bennett et al., Reference Bennett, Coleman, Tsui, Pittard and Devine2004). These studies have shed light on the selective constraints experienced by TEs, but, to date, the only empirical investigations of the effects of breeding systems on TE dynamics have focused on closely related self- and cross-pollinating plant species (Wright et al., Reference Wright, Hien Le, Schoen and Bureau2001; Tam et al., Reference Tam, Causse, Garchery, Burck, Mhiri and Granbastien2007). Both of these studies found a significant effect of the breeding system – insertion polymorphism levels were lower and individual elements segregated at higher frequencies in self-fertilizing species. Whether these conclusions apply to TEs in selfing populations more generally remains to be seen.
In the present study, we perform the first systematic survey of TE frequencies in two species of the nematode genus Caenorhabditis – the model organism Caenorhabditis elegans, which reproduces primarily as a self-fertilizing hermaphrodite, and the related outcrossing species, Caenorhabditis remanei – and test the role of breeding systems in driving TE dynamics. TEs make up about 12% of the C. elegans genome, with the most active and best characterized being type II DNA transposons of the Tc1/mariner superfamily (see the review by Bessereau, Reference Bessereau2006). These elements, which are among the most widespread DNA transposons (Plasterk et al., Reference Plasterk, Izsvák and Ivics1999), were named after the superfamily's two best-studied members: Tc1, the first TE identified in C. elegans (Emmons et al., Reference Emmons, Yesner, Ruan and Katzenberg1983; Liao et al., Reference Liao, Rosenzweig and Hirsh1983), and mariner, which was first described in Drosophila mauritiana (Jacobson et al., Reference Jacobson, Medhora and Hartl1986). These transposons have highly diverged primary sequences, but all Tc1/mariner elements probably derive from a common ancestor, and they share many common features including flanking terminal inverted repeats (TIRs) and similar modes of transposition (Plasterk et al., Reference Plasterk, Izsvák and Ivics1999). The genome of the canonical C. elegans strain N2 contains 32 copies of Tc1 (Fischer et al., Reference Fischer, Wienholds and Plasterk2003), but the copy number is strain-specific, and different wild strains harbour different numbers of Tc1 transposons, ranging from around 30 copies in most strains to upwards of 300 copies in others (Emmons et al., Reference Emmons, Yesner, Ruan and Katzenberg1983; Liao et al., Reference Liao, Rosenzweig and Hirsh1983; Egilmez et al., Reference Egilmez, Ebert and Shmookler Reis1995; Hodgkin & Doniach, Reference Hodgkin and Doniach1997). Although these elements do not show germline transposition in N2, they can be activated in the germline by mutation of a single gene in the N2 strain (Collins et al., Reference Collins, Saari and Anderson1987; Ketting et al., Reference Ketting, Haverkamp, van Luenen and Plasterk1999), activity can arise spontaneously in a non-mutator strain (Babity et al., Reference Babity, Starr and Rose1990), and some natural isolates show high rates of germline activity (Emmons et al., Reference Emmons, Yesner, Ruan and Katzenberg1983; Eide & Anderson, Reference Eide and Anderson1985).
The C. remanei genome is much more poorly characterized than that of C. elegans and, as such, much less is known about its TEs. Nonetheless, sequences homologous to Tc1 transposons are widely distributed among nematodes (Moerman & Waterston, Reference Moerman, Waterston, Berg and Howe1989), polymorphic Tc1-like elements have been found segregating in other nematode species (Hoekstra et al., Reference Hoekstra, Otsen, Lenstra and Roos1999), and probes designed for the C. elegans Tc1, Tc2, and Tc3 elements hybridize weakly in C. remanei (Abad et al., Reference Abad, Quiles, Tares, Piotte, Castagnone-Sereno, Abadon and Dalmasso1991). In addition, the associated species Caenorhabditis briggsae, which is more closely related to C. remanei than C. elegans, contains transposons similar to Tc1, and these show polymorphic hybridization patterns (Harris et al., Reference Harris, Prasad and Rose1990). Here, we describe a class of Tc1-like transposons in C. remanei, and examine both C. elegans and C. remanei for transposon population frequencies – the degree to which individual transposons segregate in natural populations. We show that levels of insertion polymorphism significantly differ between the two species and evaluate the possible role of mating system in driving TE dynamics.
2. Materials and methods
(i) Transposable elements
The canonical N2 strain of C. elegans contains 32 copies of the Tc1 DNA transposon (Fischer et al., Reference Fischer, Wienholds and Plasterk2003). We quantified the population frequencies for each of these Tc1 elements in a global sample of C. elegans strains. Transposon locations were taken from the genomic positions identified by Fischer et al. (Reference Fischer, Wienholds and Plasterk2003). For C. remanei, we identified a homologous class of transposons by using RepeatMasker (http://www.repeatmasker.org) and the Tc1 and C. briggsae Tcb1 sequences as the basis for Blastn queries of the C. remanei genome draft 15.0.1 for strain PB4641, which was sequenced using plasmid and fosmid libraries to a depth of 9·2× and assembled using the PCAP whole-genome assembly program (Washington University Genome Sequencing Center). The transposons we describe are similar to Tc1 and Tcb1 in the terminal regions, but they are smaller and may not be autonomous, so we have called them mTcre1 elements, where ‘m’ stands for miniature (N. Jiang, personal communication) and ‘re’ is an abbreviation for C. remanei. We determined the consensus sequence for mTcre1 transposons using BioEdit, and measured the population frequencies for a random subset of ‘full-length’ elements.
(ii) Nematode populations and molecular methods
For C. elegans, we tested for the presence or absence of all 32 of the Tc1 elements found in the N2 strain for each of 39 C. elegans strains, defined as isohermaphrodite lines from which we obtain single haplotypes (Table 1). Thirty-one strains were chosen to provide worldwide representation of geographic locations where this species has been found, and an additional eight strains were selected from a single sampling locality in Hermanville, France. Genetic diversity levels at nuclear loci in Hermanville samples are similar to those found on a global level (Barrière & Félix, Reference Barrière and Félix2005; Cutter, Reference Cutter2006). For C. remanei, we quantified the presence or absence for 16 elements identified in PB4641 for the inbred strain SB146 and for 14 isofemale strains from Wright State University Biological Preserve in Dayton, OH, USA (Table 1). We selected the strains from a single geographic locality for C. remanei because this population of C. remanei does not show deviation from demographic equilibrium, and so this sample is likely to be representative of the species (Cutter et al., Reference Cutter, Baird and Charlesworth2006; Cutter, Reference Cutter2008).
Table 1. C. elegans and C. remanei strains used in the present study
CGC, Caenorhabditis Genetics Center; MAF, Marie-Anne Félix; ED, Elie Dolgin; SB, Scott Baird.
For C. elegans, genomic DNA was isolated from whole plates of worms with the Puregene DNA Purification kit (D-7000A, Gentra Systems). For C. remanei, we performed whole-genome amplification of single males using the REPLI-g Mini kit (Qiagen). Diluted aliquots of the DNA samples were then used as templates for standard PCR reactions. We used DNA from the sequenced strains of C. elegans (N2) and C. remanei (PB4641) as positive controls in our PCR amplification of Tc1 and mTcre1 elements, respectively.
(iii) Population frequency assays
We determined the presence or absence of individual transposon insertions in all strains using pairs of PCR primers in the flanking regions on either side of the transposon insertion sites, coupled with a third primer matching an internal portion of the transposon sequence (e.g. Bartolomé & Maside, Reference Bartolomé and Maside2004). The PCRs performed with flanking primers produce unique sized bands that show whether the transposon is present or absent, and the reactions with the internal primers serve as additional tests to confirm transposon presence. For C. elegans, a single internal primer was used for a conserved region in 31 of the 32 Tc1 insertion sites, while a different internal primer was used for clone C50H2, due to a 701-bp deletion in the transposon (Fischer et al., Reference Fischer, Wienholds and Plasterk2003). For C. remanei, PCR failure rates were higher, presumably due to greater levels of nucleotide polymorphism, so we used two sets of flanking primers and unique internal primers for most transposon insertion sites. We used different combinations of primers until every strain was successfully amplified. Primer sets were designed for 21 mTcre1 insertions in PB4641, but for five of these, only the positive control (PB4641) worked for any primer combination. Consequently, we restrict our analysis to the 16 mTcre1 insertion sites that yielded interpretable data. The primer sequences are provided in supplementary Table S1.
Due to high levels of somatic transposition (Emmons & Yesner, Reference Emmons and Yesner1984), we could not differentiate homozygotes and heterozygotes when an insertion was detected. As a result, transposon presence functioned as a dominant marker. For C. elegans, we calculated the population frequencies by assuming that all transposons are homozygous, owing to the high degree of selfing. Therefore, the frequency of each transposon is simply equal to the number of strains for which the insertion was detected, divided by the total number of strains (excluding N2). For C. remanei, we first discriminated between X-linked vs. autosomal insertions by inferring the chromosomal location of each insertion using Blastn of the unique flanking regions against the 2005 preliminary assembly of the C. remanei genome in Wormbase (http://www.wormbase.org). Based on wobble-aware bulk aligner (WABA) alignments of the resulting C. remanei contigs to the C. elegans genome, we then inferred the likely syntenic chromosome. Because we used single male DNA preparations for C. remanei, heterozygosity of X-linked insertions is not possible, so the frequency of such mTcre1 insertions is simply the observed population frequency. For autosomal insertions, we assumed random mating, which seems to be appropriate for the Ohio sample (Cutter et al., Reference Cutter, Baird and Charlesworth2006; Cutter, Reference Cutter2008), and calculated the frequency of each transposon according to Hardy–Weinberg expectations as one minus the square root of the frequency of strains that lacked the insertion. Frequency calculations were restricted to the 14 Ohio samples, with the inbred SB146 strain excluded from the analysis.
(iv) Estimating the strength of selection
We used the diffusion approximation methods derived by Petrov et al. (Reference Petrov, Aminetzrach, Davis, Bensasson and Hirsh2003) to estimate the probability that an insertion is at a particular population frequency, and calculated a maximum likelihood estimate of the selection coefficient acting on the transposons. We calculated 95% confidence intervals around the maximum likelihood scores to obtain measures of the intensity of natural selection, N es, assuming semi-dominance (i.e. h=0·5), effective population sizes of 104 and 106 for C. elegans and C. remanei, respectively (Cutter, Reference Cutter2006, 2008; Cutter et al., Reference Cutter, Baird and Charlesworth2006), and that all transposons have independent effects and are subject to the same strength of selection, s. Qualitative conclusions were unaffected by increasing or decreasing the effective population sizes one order of magnitude (results not shown). This analysis only considers segregating elements (i.e. not fixed) and, by using posterior probability functions, accounts for the fact that, by only studying insertions present in the reference sequenced strain, we have pre-sampled transposons in proportion to their population frequencies (Petrov et al., Reference Petrov, Aminetzrach, Davis, Bensasson and Hirsh2003). For C. elegans, the analysis was done for the entire collection of 39 strains, for the 8 strains from Hermanville on their own, and for 30 random ‘scattered sample’ subsets with a single strain selected from each geographic location (23 strains in total). This subsampling approach of taking a single strain from each locality may approximate a homogeneously mixing population for a large number of localities connected by migration (Wakeley, Reference Wakeley2003; Cutter, Reference Cutter2006; Matsen & Wakeley, Reference Matsen and Wakeley2006). For C. remanei, the analysis was limited to the Ohio population.
This analysis assumes a large number of independently segregating insertions at transposition–selection balance. To test this, we measured the level of neutral nucleotide diversity among Tc1 transposons within the genome of the canonical N2 strain and total nucleotide diversity among ‘full-length’ mTcre1 transposons within the PB4641 C. remanei genome using DnaSP 4.10.9 (Rozas et al., Reference Rozas, Sanchez-DelBarrio, Messeguer and Rozas2003). At copy number equilibrium, population genetic theory predicts that the level of neutral nucleotide diversity among active transposons is equal to 4N eΛμ, where μ is the neutral mutation rate, Λ is the average number of active transposons per haploid genome and 2N eΛ is the effective population size of the transposon family (Brookfield, Reference Brookfield1986; Sánchez-Garcia et al., Reference Sánchez-Garcia, Maside and Charlesworth2005). Therefore, silent site diversity between transposons is expected to equal the average haploid copy number multiplied by the average genomic silent-site diversity, assuming that the mutation rate in transposons is similar to that of the genome as a whole (Brookfield, Reference Brookfield1986). We also measured levels of linkage disequilibrium among transposons for the 39 C. elegans strains (excluding N2) and for the 14 Ohio C. remanei strains, using the squared correlation between pairs of sites (r 2). Because our sample size, n, for each species is small, the effective population size multiplied by the recombination rate is expected to be much greater than the sample size, and the expected linkage disequilibrium, E(r 2)≈1/n (Weir & Hill, Reference Weir and Hill1980). Therefore, we subtract 1/n from our r 2-values, and compare deviations from expectations for the two species.
With the well-annotated genome of C. elegans, we also tested for correlations between the population frequency of Tc1 transposons and various aspects of the genomic environment: local recombination rate, gene density, transposon polymorphism levels [single nucleotide polymorphism (SNPs) and indels], and whether the transposon is inserted in an intron. Recombination rates based on the nearest ten loci were taken from Cutter & Payseur (Reference Cutter and Payseur2003), the numbers of SNPs and indels were obtained from Fischer et al. (Reference Fischer, Wienholds and Plasterk2003), and gene density estimates and whether the transposon was found in an intron were determined using Wormbase (http://www.wormbase.org).
3. Results
(i) C. remanei transposons
We identified a class of transposons, which we refer to as mTcre1 transposons, exhibiting high sequence similarity to the ends of the TIRs of Tc1 and Tcb1 elements. Figure 1 shows the consensus mTcre1 sequence, which matches 17 of the outer 18 bp in the TIR of Tc1, and 30 of the outer 31 bp in the Tcb1 sequence. The mTcre1 elements have longer TIRs and a shorter total length than either Tc1 or Tcb1. The unique 86 bp internal sequence is probably too short to contain an open reading frame; however, the conserved portion of the TIRs probably contains a transposase binding site, because the Tc1 transposase binding site was identified within the outer portion of the Tc1 inverted repeats (Vos & Plasterk, Reference Vos and Plasterk1994). In this way, mTcre1 elements might more closely resemble other Tc-family transposons such as C. elegans Tc7 elements, which are shorter than Tc1 and rely on Tc1-derived transposase activity (Rezsohazy et al., Reference Rezsohazy, van Luenen, Durbin and Plasterk1997). Furthermore, mTcre1 elements are flanked by TA dinucleotides, just like Tc1 and Tcb1, which presumably result from target site sequence duplication upon integration (van Luenen et al., Reference van Luenen, Colloms and Plasterk1994). We identified 81 mTcre1 transposons in total, inferred from similarity to the TIRs, although this is probably an underestimate of the true number of mTcre1 elements in the PB4641 genome. Of these, 80% are ‘full-length’ (i.e. similar in length to the consensus mTcre1 sequence), 11% contain large (>100 bp) deletions and 9% have complex insertions making them at least 400 bp larger.
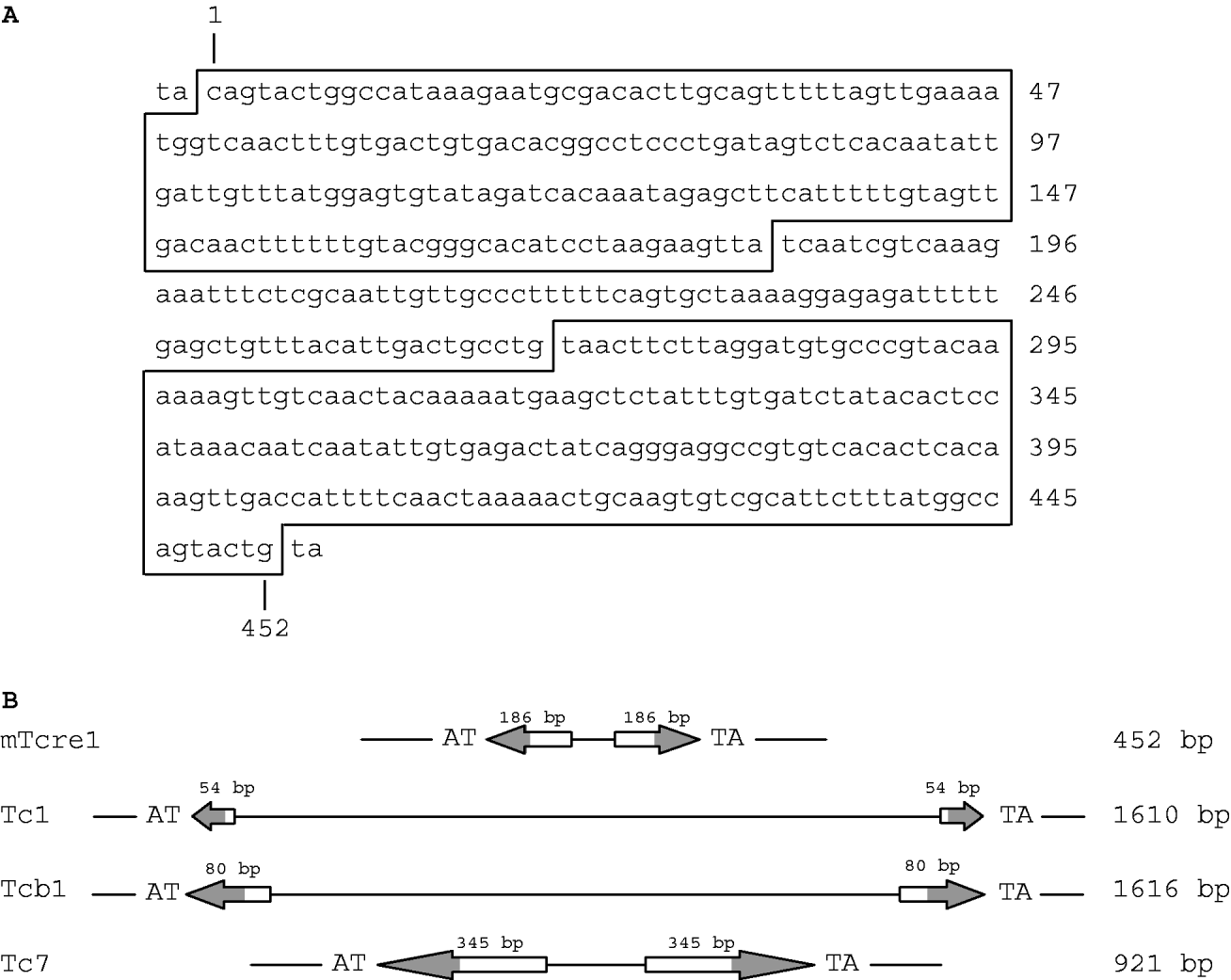
Fig. 1. (A) Consensus nucleotide sequence of the mTcre1 C. remanei transposon, with the TIRs shown in boxes. (B) Schematic representation of mTcre1 showing homology to Tc1, Tcb1 and Tc7. Arrows represent the TIRs, with the shaded areas corresponding to regions of sequence similarity.
(ii) Transposon population frequencies
Whether each transposon was present or absent in all the strains tested is shown in Fig. 2. Individual C. elegans strains were significantly more likely than C. remanei strains to harbour the transposons tested (Mann–Whitney U=568·5, P<10−6); on average, ~80% of the 32 Tc1 transposons found in N2 were present in any given wild strain of C. elegans, while C. remanei strains only had around half of the mTcre1 elements detected in PB4641. The population frequencies of transposons were also much higher for Tc1 elements in C. elegans than for mTcre1 elements in C. remanei (Mann–Whitney U=454·5, P<10−6), as the frequency spectrum was skewed towards high-frequency elements in C. elegans (Fig. 3). A greater proportion of insertions was fixed for Tc1 (9 of 32 elements) than for mTcre1 (1 of 16), although this difference was not statistically significant (Fisher's Exact Test, P=0·13). For C. elegans, the population frequency of Tc1 transposons did not correlate with recombination rate, gene density, polymorphism levels or whether the transposon was found in an intron (all Spearman's |ρ|<0·20, P>0·30). The presence/absence status of transposons also showed no strong signature of geographic structuring in C. elegans, consistent with other studies of genetic diversity (Denver et al., Reference Denver, Morris and Thomas2003; Barrière & Félix, Reference Barrière and Félix2005; Haber et al., Reference Haber, Schüngel, Putz, Müller, Hasert and Schulenburg2005; Cutter, Reference Cutter2006; Dolgin et al., Reference Dolgin, Félix and Cutter2008).
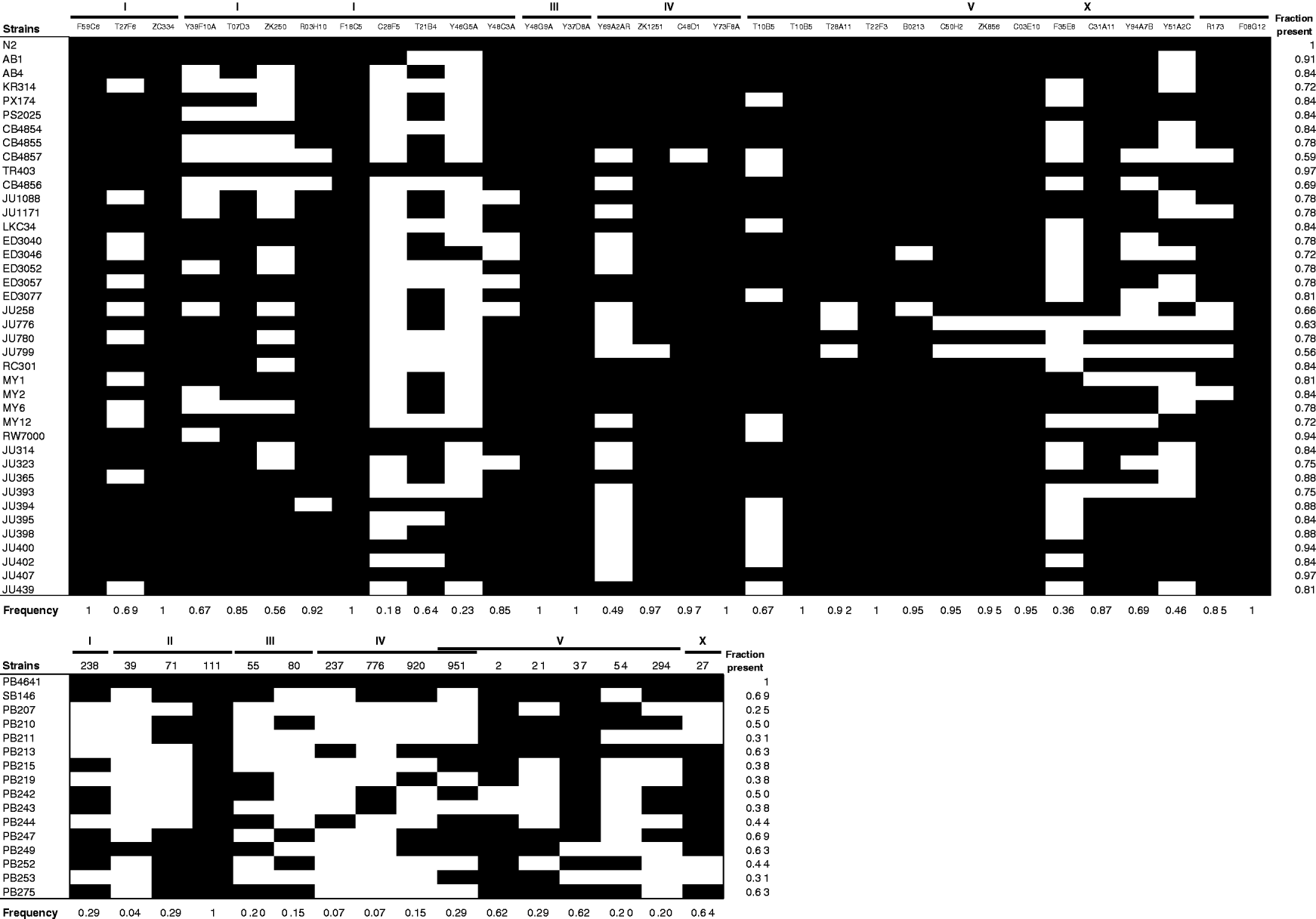
Fig. 2. Transposon frequencies for C. elegans (A) and C. remanei (B). Dark squares indicate presence and white squares indicate absence of the transposon. Population frequencies of each transposon calculated for the 39 C. elegans strains (excluding N2) and for the 14 Ohio C. remanei strains are shown along the bottom; the fraction of elements found for each strain is shown along the right; and the position of each transposon, denoted by the clone (C. elegans) or supercontig (C. remanei) and the linkage group, is shown along the top. For C. remanei supercontig 951, the WABA alignment was inconclusive between linkage groups IV and V.
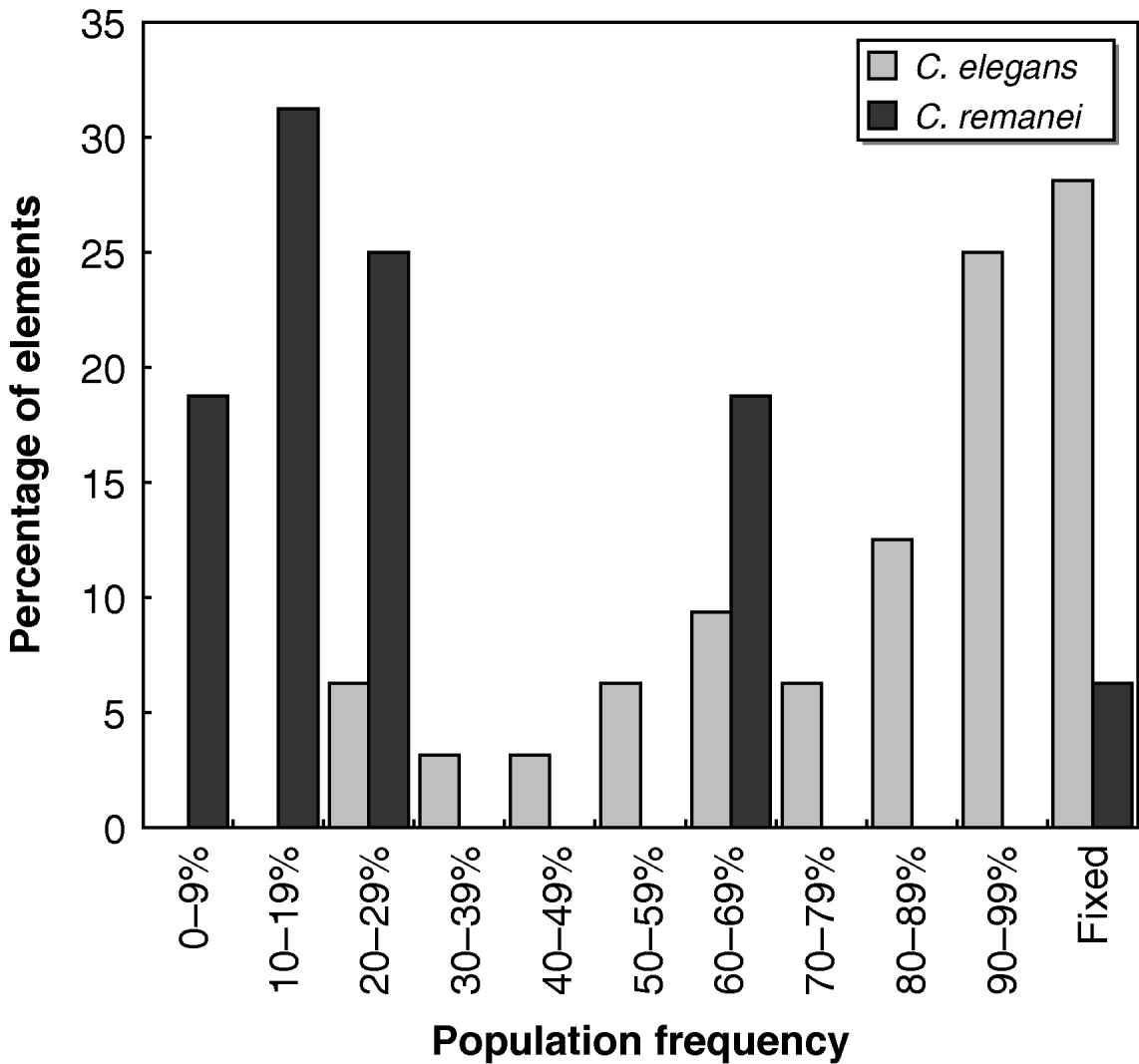
Fig. 3. Histogram of population frequencies of C. elegans Tc1 transposons (light shaded bars) and C. remanei mTcre1 transposons (dark shaded bars).
(iii) Strength of selection
Using the observed population frequency distributions of polymorphic transposons to estimate the maximum likelihood strength of selection, we found evidence for purifying selection (N es<0) in C. remanei but not in C. elegans. For mTcre1 elements in C. remanei, N es=−2·2 (95% confidence interval: −3·9⩽N es⩽−0·6). In contrast, when considering all C. elegans strains tested for Tc1 elements, N es=5·9 (1·3⩽N es⩽27·3), suggesting that Tc1 elements are subject to positive selection in C. elegans. This seems unlikely, however, because Tc1 transposons cause large mutagenic effects when transpositionally active in the germline (Plasterk & van Luenen, Reference Rezsohazy, van Luenen, Durbin and Plasterk1997; Bégin & Schoen, Reference Bégin and Schoen2006, Reference Barrière and Félix2007).
The positive N es value for C. elegans arises from the large number of high-frequency transposons (see Fig. 3), which could be an artefact of population structure if many strains shared a recent N2-like common ancestor. To attempt to remove the effects of population subdivision, we created random subsets of ‘scattered samples’ with only a single strain from each geographic locality (Wakeley, Reference Wakeley2003; Cutter, Reference Cutter2006; Matsen & Wakeley, Reference Matsen and Wakeley2006). This analysis gave an average strength of selection, N es≈4. However, heterogeneity between strains from the same location led to 95% confidence intervals for different scattered random subsets to imply no significant difference from neutrality in 7 of 30 subsets, and positive selection in the remaining 23. Overall, the range of lower 95% confidence intervals for the 30 random scattered subsamples spanned the range −0·2 to 1·1. Considering just the eight strains from a single locality in Hermanville, France, we found no significant difference from neutrality: N es=1·5 (−1·1⩽N es⩽16·8). Further, the lack of evidence for purifying selection in C. elegans is not an artefact of the assumption of complete homozygosity. If we assume Hardy–Weinberg ratios of allele frequencies, as we did for C. remanei, we still observe no significant departure from neutral expectations when considering all 39 strains: N es=0·3 (−1·1⩽N es⩽2·8).
The maximum likelihood method for calculating the strength of selection assumes that transposons are at copy number equilibrium. To evaluate this assumption, we contrasted predictions of nucleotide diversity and linkage disequilibrium with observed values (see the ‘Materials and Methods’ section). For C. remanei, total nucleotide diversity among ‘full-length’ mTcre1 elements in the PB4641 genome was π=18·2% and θ=21·3%. These diversity measures include parts of the transposon that might experience selection, including the inverted repeats and conserved region, so silent site diversity is presumably even greater. Nonetheless, these values are much greater than the 3·6–4·7% overall silent-site diversity for C. remanei (Cutter et al., Reference Cutter, Baird and Charlesworth2006; Cutter, Reference Cutter2008), consistent with equilibrium expectations (Brookfield, Reference Brookfield1986). In contrast, we found that nucleotide diversity at silent sites among the 32 C. elegans Tc1 elements in the N2 genome was πsi=0·19% and θsi=0·76%. These values are similar to the overall silent-site diversity of 0·2–0·3% (Cutter, Reference Cutter2006), and not ~30× greater as predicted by equilibrium theory (Brookfield, Reference Brookfield1986). Furthermore, this analysis may be conservative as extra rounds of DNA replication during transposition could enhance the nucleotide mutation rate of TEs above that of the rest of the host's genome. We also found Tajima's D=−1·89 (P<0·05), indicating an excess of low-frequency variants among Tc1 elements, and the mean deviation of linkage disequilibrium from expectation was greater for C. elegans: 0·056 between all pairs of sites and 0·112 for intrachromosomal comparisons, compared with r 2-deviations for C. remanei of 0·025 and 0·010, respectively. A greater proportion of r 2-values was also significant by Fisher's exact tests in C. elegans than C. remanei (Fisher's exact test between species, P=0·002). Together, these measures all suggest that the assumption of copy number equilibrium might be violated for C. elegans.
4. Discussion
Phylogenetic evidence indicates that the ancestor of C. elegans was obligately outcrossing with separate male and female individuals (Kiontke & Fitch, Reference Kiontke and Fitch2005). Thus, C. remanei provides a useful proxy for the ancestral state of C. elegans, and comparisons between the two species provide a way of detecting changes in the evolutionary pressures experienced by C. elegans upon adopting a self-fertilizing breeding system. Analyses of sequence polymorphism show that C. elegans has much lower levels of genetic variation than C. remanei (Graustein et al., Reference Graustein, Gaspar, Walters and Palopoli2002; Jovelin et al., Reference Jovelin, Ajie and Phillips2003; Haag & Ackerman, Reference Haag and Ackerman2005; Cutter, Reference Cutter2006, Reference Cutter and Payseur2008; Cutter et al., Reference Cutter, Baird and Charlesworth2006). C. remanei also suffers from much stronger inbreeding depression than C. elegans (Dolgin et al., Reference Dolgin, Charlesworth, Baird and Cutter2007), and the two species display very different mating behaviours (Chasnov et al., Reference Chasnov, So, Chan and Chow2007; Garcia et al., Reference Garcia, LeBoeuf and Koo2007). Here, we show that natural populations of C. elegans and C. remanei also show markedly different transposon frequency distributions. The greater proportion of polymorphic insertions in C. remanei segregating at lower population frequencies compared with C. elegans is consistent with an important role of breeding systems contributing to the control of transposon dynamics in natural populations, and with other comparative studies of TEs in the plant genera Arabidopsis (Wright et al., Reference Wright, Hien Le, Schoen and Bureau2001) and Solanum (Tam et al., Reference Tam, Causse, Garchery, Burck, Mhiri and Granbastien2007).
The estimates of N es reveal distinctly different intensities of natural selection against transposons in C. elegans and C. remanei. We found signs of purifying selection against element mTcre1 insertions in C. remanei, as indicated by negative N es values, but no such evidence for Tc1 elements in C. elegans. It should be noted, however, that the elements examined in each species are not strictly homologous, which could potentially influence the contrast of TE polymorphism patterns between the two species. Since the unique internal sequence of mTcre1 elements probably does not contain an open reading frame, these transposons are probably not autonomous and might experience different dynamics from the autonomous Tc1 elements. However, one would expect non-autonomous elements to be less mobile, and thus more likely to be fixed or at high frequencies (Bartolomé & Maside, Reference Bartolomé and Maside2004), whereas we see the opposite result. This potential innate difference between the elements would make our species contrast conservative. Alternatively, selection for self-regulation in selfing lineages could drive lower transposition rates in C. elegans (Charlesworth & Langley, Reference Charlesworth and Langley1986), or the effective transposition rate might be lower in C. elegans if transposons cannot easily invade and spread into different genetic backgrounds with self-fertilization. If transposition rates are lower for Tc1 elements than for mTcre1 elements, fewer elements would be of recent origin in C. elegans, and population frequencies could be skewed upwards. In C. remanei, on the other hand, if selection effectively removes most elements, we might expect only to find either newly transposed elements at low frequencies or to observe old insertions that achieved high frequencies by drift. This would be consistent with the somewhat bimodal distribution for mTcre1 elements seen in Fig. 3.
At first sight, our analysis of the intensity of selection suggests neutrality or positive selection for Tc1 elements in C. elegans. Although TEs in general can sometimes be beneficial (Kidwell & Lisch, Reference Kidwell and Lisch2001; Schlenke & Begun, Reference Schlenke and Begun2004), this is unlikely to be generally true for Tc1 elements, which are known to cause strong deleterious mutational effects (Plasterk & van Luenen, Reference Plasterk, van Luenen, Riddle, Blumenthal, Meyer and Priess1997; Bégin & Schoen, Reference Bégin and Schoen2006, Reference Bégin and Schoen2007). What might bias N es estimates for C. elegans? One possibility is violation of the assumption of copy number equilibrium required by the maximum likelihood method used for calculating the intensity of selection, as estimates of nucleotide diversity are lower than equilibrium predictions. However, the low estimates of genetic diversity between insertions in N2 could potentially result from gene conversion among Tc1 elements, thus biasing the molecular diversity estimates. The observation that Tc1 elements can acquire the sequence of other Tc1 elements elsewhere in the genome suggests that there might be continuous exchange of sequence information between individual insertions (Fischer et al., Reference Fischer, Wienholds and Plasterk2003). But if between elements gene conversion is unbiased, theory shows that this should not affect diversity measures (Ohta, Reference Ohta1985; Slatkin, Reference Slatkin1985). We cannot exclude the possibility that gene conversion is biased, which could reduce diversity. However, there is no reason why this should cause the excess of low-frequency variants that we detected as a negative Tajima's D, because gene conversion would introduce variants randomly across the genealogy (Marais, Reference Marais2003).
The greater proportion of sites in linkage disequilibrium in C. elegans also suggests that the assumption of independence between insertions might be violated; although it is interesting to note that the levels of linkage disequilibrium reported here between TEs are much lower than the estimates of linkage disequilibrium in C. elegans from sequence data (Cutter, Reference Cutter2006, Reference Cutter2008; Cutter et al., Reference Cutter, Baird and Charlesworth2006) or other molecular markers (Barrière & Félix, Reference Barrière and Félix2005, Reference Barrière and Félix2007; Haber et al., Reference Haber, Schüngel, Putz, Müller, Hasert and Schulenburg2005). This could reflect a difference in timescale between transposons and other mutational processes. If transposition rates greatly exceed mutation rates, then transposon insertions will tend to be more recent, making them more likely to be independent of each other. This idea is supported by the significantly negative value for Tajima's D, which is also consistent with a recent burst of transposition (Sánchez-Garcia et al., Reference Sánchez-Garcia, Maside and Charlesworth2005). However, this would then imply that strains lacking germline Tc1 activity only recently acquired suppressors of transposition (Collins et al., Reference Collins, Saari and Anderson1987; Babity et al., Reference Babity, Starr and Rose1990; Mori et al., Reference Mori, Moerman and Waterston1990), and the presence of numerous fixed and high-frequency transposons indicates that many insertions preceded strain divergence. If we compare silent-site diversity among Tc1 elements from the N2 strain between fixed and near-fixed (⩾95% population frequency) insertions with polymorphic insertions, we observe a nearly two-fold difference in diversity levels (fixed/near-fixed: πsi=0·31% and θsi=0·73%; polymorphic: πsi=0·17% and θsi=0·41%), further suggesting that the high-frequency elements are more ancient. One explanation for this pattern is that there was a period of transpositional activity in the past, with the resulting insertions drifting to high frequencies or fixation, and the lower frequency TEs seen in our study represent a more recent period of activity. This resembles the pattern seen in Drosophila melanogaster (Bartolomé & Maside, Reference Bartolomé and Maside2004).
One scenario to explain the high number of fixed or near-fixed insertions across all chromosomes in C. elegans is that a nearly genome-wide selective sweep of an N2-like genotype with low levels of excision occurred in a number of strains. Such an event, however, would be unlikely to account for around half the insertions that show intermediate frequencies. Further, the complete lack of low-frequency (<20%) insertions in C. elegans, compared with around half the elements with low frequencies in C. remanei (see Fig. 3), suggests that the observed pattern is more likely to reflect differences in selection pressures between the species. Therefore, we caution against drawing strong conclusions from the positive estimates of selection for C. elegans from this analysis, because of the likelihood that population demographic processes might skew the transposon frequency spectrum. Rather, we argue that segregating Tc1 elements are probably selectively neutral, as seen in the analysis of the Hermanville population and some of the scattered samples.
Two main factors may be involved in causing the observed differences in transposon profiles. First, the skewed distribution in C. elegans towards high-frequency elements suggests a reduction in the efficacy of purifying selection in selfing lineages, due to a smaller effective population size (Wright & Schoen, Reference Wright and Schoen1999; Morgan, Reference Morgan2001). New transposons may persist longer in a polymorphic state in C. remanei and selection should be more efficient at eliminating them, because its effective population size is estimated to be two orders of magnitude larger than that of C. elegans (Cutter, Reference Cutter2006, Reference Cutter2008; Cutter et al., Reference Cutter, Baird and Charlesworth2006). Assuming that transposons have similar selective effects in both species, such large population size differences could explain the different N es estimates. But transposons could experience different selective regimes due to differences in levels of homozygosity between the breeding systems. When homozygous and heterozygous insertions have distinct fitness effects, changes in selfing rate can have dramatic effects on TE dynamics (Wright & Schoen, Reference Wright and Schoen1999; Morgan, Reference Morgan2001). Under the ectopic exchange model, selection against homozygous insertions is expected to be weak or null, whereas under the deleterious insertion model, selection will be strongest against homozygous insertions (see the review by Nuzhdin, Reference Nuzhdin1999). Heterozygosity levels in C. elegans are typically very low (Barrière & Félix, Reference Barrière and Félix2005, Reference Barrière and Félix2007; but see Sivasundar & Hey, Reference Sivasundar and Hey2005), suggesting that most insertions will be in a homozygous state, which would reduce the opportunity for ectopic pairing (Montgomery et al., Reference Montgomery, Huang, Langley and Judd1991). Therefore, a second explanation for the high population frequencies of elements in C. elegans is that selection is weaker against the greater proportion of homozygous insertions, as predicted by the ectopic exchange model.
Previous studies of polymorphic Tc1 elements in C. elegans have compared the canonical N2 strains with only a limited number of natural isolates (Emmons et al., Reference Emmons, Yesner, Ruan and Katzenberg1983; Liao et al., Reference Liao, Rosenzweig and Hirsh1983; Eide & Anderson, Reference Eide and Anderson1985; Harris & Rose, Reference Harris and Rose1989; Egilmez et al., Reference Egilmez, Ebert and Shmookler Reis1995; Hodgkin & Doniach, Reference Hodgkin and Doniach1997). Here, we studied C. elegans strains representing a complete global sampling covering all six major continents where C. elegans has been found, as well as a number of strains derived from a single geographic locality. Nevertheless, we observe many similarities in our dataset to those observed earlier for only a handful of strains. Egilmez et al. (Reference Egilmez, Ebert and Shmookler Reis1995) assessed Tc1-site occupancy among five strains, including N2, and found that 20 of the 32 Tc1 insertions in N2 were common to all five strains. With our larger sample of 40 strains, we also observed high numbers of fixed insertions (9 of 32). Following the observation that the Bergerac strain had active germline transposition and many times more Tc1 elements than N2 (Emmons et al., Reference Emmons, Yesner, Ruan and Katzenberg1983; Liao et al., Reference Liao, Rosenzweig and Hirsh1983), much of the focus on describing natural variation in Tc1 elements has been on characterizing strains as either ‘low-copy’ (~30 elements) or ‘high-copy’ (>300 elements) strains. We have not quantified copy numbers in the present study, and not all the strains used have been characterized previously for transposon abundance, but it is interesting to note that the two high-copy strains identified previously, RW7000/Bergerac and TR403 (Egilmez et al., Reference Egilmez, Ebert and Shmookler Reis1995; Hodgkin & Doniach, Reference Hodgkin and Doniach1997), were also among the strains containing the highest fraction of insertions present (see Fig. 2 A). This confirms previous findings that nearly all of the Tc1 elements in N2 are in the same location in the high-copy Bergerac strain (Harris & Rose, Reference Harris and Rose1989).
An outstanding question that cannot be answered by our method is how the total numbers of transposons in natural isolates compare within and between the two species. In C. elegans, Tc1 copy number ranges an order of magnitude between wild strains (Egilmez et al., Reference Egilmez, Ebert and Shmookler Reis1995; Hodgkin & Doniach, Reference Hodgkin and Doniach1997). In a single C. remanei genome (PB4641), we identified 81 mTcre1 elements by bioinformatics approaches, although this is likely to underestimate the true copy number, and we do not know how copy numbers vary among strains. Furthermore, it is unclear how biologically relevant it is to compare the total genomic abundance of Tc1 and mTcre1 elements if they are not homologous. Previous studies in related self- and cross-pollinating plant species similarly found that outcrossing species consistently showed lower population frequencies of elements (Wright et al., Reference Wright, Hien Le, Schoen and Bureau2001; Tam et al., Reference Tam, Causse, Garchery, Burck, Mhiri and Granbastien2007), but not of element copy number. Whereas self-fertilizing Arabidopsis had slightly higher copy numbers of DNA transposons than outcrossing species (Wright et al., Reference Wright, Hien Le, Schoen and Bureau2001), no relationship was found between retrotransposon number and breeding system among related tomato species (Tam et al., Reference Tam, Causse, Garchery, Burck, Mhiri and Granbastien2007). Additional comparisons of TEs in other closely related species or using other TE families should help illuminate the effect of breeding system on total abundance. For Caenorhabditis, more work is needed to characterize and quantify transposons in C. remanei in particular.
The results presented here support a role of breeding systems in driving TE dynamics. Two independent derivations of self-fertilization in Caenorhabditis (Kiontke & Fitch, Reference Kiontke and Fitch2005) among the six species with sequenced genomes will provide a useful platform for testing the generality of breeding system as an important factor in TE evolution. Future studies will help to further tease apart the selection pressures imposed by TEs, and the impact of selfing rates on TE dynamics.
We are grateful to D. Petrov for sharing Mathematica files for the selection analysis, and to A. Betancourt and S. Wright for helpful comments on the manuscript. Worm strains were generously provided by S. Baird, M.-A. Félix and the Caenorhabditis Genetics Center. This work was funded by a Scottish International Education Trust travel grant, and by postgraduate scholarships from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the University of Edinburgh School of Biological Sciences to E. S. D., and by an NSERC Discovery Grant to A. D. C. B. C. was supported by the Royal Society (UK).








